Abstract
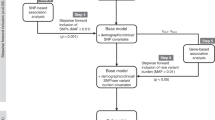
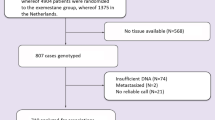
The dose of docetaxel is currently calculated based on body surface area and does not reflect the pharmacokinetic, metabolic potential or genetic background of the patients. The influence of genetic variation on the clearance of docetaxel was analysed in a two-stage analysis. In step one, 583 single-nucleotide polymorphisms (SNPs) in 203 genes were genotyped on samples from 24 patients with locally advanced non-small cell lung cancer. We found that many of the genes harbour several SNPs associated with clearance of docetaxel. Most notably these were four SNPs in EGF, three SNPs in PRDX4 and XPC, and two SNPs in GSTA4, TGFBR2, TNFAIP2, BCL2, DPYD and EGFR. The multiple SNPs per gene suggested the existence of common haplotypes associated with clearance. These were confirmed with detailed haplotype analysis. On the basis of analysis of variance (ANOVA), quantitative mutual information score (QMIS) and Kruskal–Wallis (KW) analysis SNPs significantly associated with clearance of docetaxel were confirmed for GSTA4, PRDX4, TGFBR2 and XPC and additional putative markers were found in CYP2C8, EPHX1, IGF2, IL1R2, MAPK7, NDUFB4, TGFBR3, TPMT (2 SNPs), (P<0.05 or borderline significant for all three methods, 14 SNPs in total). In step two, these 14 SNPs were genotyped in additional 9 samples and the results combined with the genotyping results from the first step. For 7 of the 14 SNPs, the results are still significant/borderline significant by all three methods: ANOVA, QMIS and KW analysis strengthening our hypothesis that they are associated with the clearance of docetaxel.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Abbreviations
- AAG:
-
α1-acid glycoprotein
- AUC:
-
area under the curve
- BSA:
-
body surface area
- HPLC:
-
high performance liquid chromatography
- KW:
-
Kruskal–Wallis
- LD:
-
linkage disequilibrium
- LOOCV:
-
leave one out cross validation
- MAF:
-
minor allele frequency
- MIS:
-
mutual information score
- NSCLC:
-
non-small cell lung cancer
- OMIM:
-
online Mendelian inheritance in man
- QMIS:
-
quantitative mutual information score
- ROS:
-
reactive oxygen species
- SNPs:
-
single-nucleotide polymorphisms
- UHT:
-
ultra high throughput
References
Morse DL, Gray H, Payne CM, Gillies RJ . Docetaxel induces cell death through mitotic catastrophe in human breast cancer cells. Mol Cancer Ther 2005; 4: 1495–1504.
Wang LG, Liu XM, Kreis W, Budman DR . The effect of antimicrotubule agents on signal transduction pathways of apoptosis: a review. Cancer Chemother Pharmacol 1999; 44: 355–361.
Baker SD, Sparreboom A, Verweij J . Clinical pharmacokinetics of docetaxel: recent developments. Clin Pharmacokinet 2006; 45: 235–252.
Caffo O . Radiosensitization with chemotherapeutic agents. Lung Cancer 2001; 34 (Suppl 4): S81–S90.
Brunsvig PF, Hatlevoll R, Berg R, Lauvvang G, Owre K, Wang M et al. Weekly docetaxel with concurrent radiotherapy in locally advanced non-small cell lung cancer: a phase I/II study with 5 years′ follow-up. Lung Cancer 2005; 50: 97–105.
Baker SD, Verweij J, Cusatis GA, van Schaik RH, Marsh S, Orwick SJ et al. Pharmacogenetic pathway analysis of docetaxel elimination. Clin Pharmacol Ther 2009; 85: 155–163.
Bruno R, Vivler N, Vergniol JC, De Phillips SL, Montay G, Sheiner LB . A population pharmacokinetic model for docetaxel (Taxotere): model building and validation. J Pharmacokinet Biopharm 1996; 24: 153–172.
Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem 2000; 275: 39907–39913.
Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997; 236: 313–322.
Venugopal R, Jaiswal AK . Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA 1996; 93: 14960–14965.
Dai Y, Rashba-Step J, Cederbaum AI . Stable expression of human cytochrome P4502E1 in HepG2 cells: characterization of catalytic activities and production of reactive oxygen intermediates. Biochemistry 1993; 32: 6928–6937.
Morel Y, Barouki R . Repression of gene expression by oxidative stress. Biochem J 1999; 342 (Pt 3): 481–496.
Raza H, John A . 4-hydroxynonenal induces mitochondrial oxidative stress, apoptosis and expression of glutathione S-transferase A4-4 and cytochrome P450 2E1 in PC12 cells. Toxicol Appl Pharmacol 2006; 216: 309–318.
Park JY, Shigenaga MK, Ames BN . Induction of cytochrome P4501A1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin or indolo(3,2-b)carbazole is associated with oxidative DNA damage. Proc Natl Acad Sci USA 1996; 93: 2322–2327.
Edvardsen H, Irene Grenaker AG, Tsalenko A, Mulcahy T, Yuryev A, Lindersson M et al. Experimental validation of data mined single nucleotide polymorphisms from several databases and consecutive dbSNP builds. Pharmacogenet Genomics 2006; 16: 207–217.
Gurney H . Dose calculation of anticancer drugs: a review of the current practice and introduction of an alternative. J Clin Oncol 1996; 14: 2590–2611.
Andersen A, Warren DJ, Brunsvig PF, Aamdal S, Kristensen GB, Olsen H . High sensitivity assays for docetaxel and paclitaxel in plasma using solid-phase extraction and high-performance liquid chromatography with UV detection. BMC Clin Pharmacol 2006; 6: 2.
Tsalenko A, Ben-Dor A, Cox N, Yakhini Z . Methods for analysis and visualization of SNP genotype data for complex diseases. Pac Symp Biocomput 2003: 548–561.
Rice JA . Mathematical Statistics and Data Analysis, 3rd edn. Australia; Belmont, CA: Thompson/Brooks/Cole, c2007.
Tsalenko A, Sharan R, Edvardsen H, Kristensen VN, Borresen-Dale AL, Ben-Dor A et al. Analysis of SNP-expression association matrices. J BioComput Bioinformat 2006; 4: 259–274.
Hedenfalk I, Ringner M, Ben-Dor A, Yakhini Z, Chen Y, Chebil G et al. Molecular classification of familial non-BRCA1/BRCA2 breast cancer. Proc Natl Acad Sci USA 2003; 100: 2532–2537.
Barrett JC, Fry B, Maller J, Daly MJ . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
McDonald SL, Stevenson DA, Moir SE, Hutcheon AW, Haites NE, Heys SD et al. Genomic changes identified by comparative genomic hybridisation in docetaxel-resistant breast cancer cell lines. Eur J Cancer 2005; 41: 1086–1094.
Kristensen VN, Edvardsen H, Tsalenko A, Nordgard SH, Sorlie T, Sharan R et al. Genetic variation in putative regulatory loci controlling gene expression in breast cancer. Proc Natl Acad Sci USA 2006; 103: 7735–7740.
Iwao-Koizumi K, Matoba R, Ueno N, Kim SJ, Ando A, Miyoshi Y et al. Prediction of docetaxel response in human breast cancer by gene expression profiling. J Clin Oncol 2005; 23: 422–431.
Lee AJ, Conney AH, Zhu BT . Human cytochrome P450 3A7 has a distinct high catalytic activity for the 16alpha-hydroxylation of estrone but not 17beta-estradiol. Cancer Res 2003; 63: 6532–6536.
Chen H, Fantel AG, Juchau MR . Catalysis of the 4-hydroxylation of retinoic acids by cyp3a7 in human fetal hepatic tissues. Drug Metab Dispos 2000; 28: 1051–1057.
Hubatsch I, Ridderstrom M, Mannervik B . Human glutathione transferase A4-4: an alpha class enzyme with high catalytic efficiency in the conjugation of 4-hydroxynonenal and other genotoxic products of lipid peroxidation. Biochem J 1998; 330: 175–179.
Wang WL, Yeh SF, Chang YI, Hsiao SF, Lian WN, Lin CH et al. PICK1, an anchoring protein that specifically targets protein kinase Calpha to mitochondria selectively upon serum stimulation in NIH 3T3 cells. J Biol Chem 2003; 278: 37705–37712.
Nagase H, Mao JH, Balmain A . Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res 2003; 63: 4849–4853.
Acknowledgements
This project was supported by the Norwegian Cancer Association (project number D-03067 and D-99061) and the National Programme for Research in Functional Genomics in Norway (FUGE, project numbers 151924/150 and 15204/150), funded by the Research Council of Norway. Genotyping was partly performed at the Uppsala SNP technology platform, which is funded by the Swedish Wallenberg Consortium North (WCN).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Edvardsen, H., Brunsvig, P., Solvang, H. et al. SNPs in genes coding for ROS metabolism and signalling in association with docetaxel clearance. Pharmacogenomics J 10, 513–523 (2010). https://doi.org/10.1038/tpj.2010.6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2010.6
Keywords
This article is cited by
-
Role of genetic variation in docetaxel-induced neutropenia and pharmacokinetics
The Pharmacogenomics Journal (2016)
-
Folate-Mediated Targeted Delivery of Combination Chemotherapeutics Loaded Reduced Graphene Oxide for Synergistic Chemo-Photothermal Therapy of Cancers
Pharmaceutical Research (2016)