Abstract
The histaminergic system (HS) has a critical role in cognition, sleep and other behaviors. Although not well studied in autism spectrum disorder (ASD), the HS is implicated in many neurological disorders, some of which share comorbidity with ASD, including Tourette syndrome (TS). Preliminary studies suggest that antagonism of histamine receptors 1–3 reduces symptoms and specific behaviors in ASD patients and relevant animal models. In addition, the HS mediates neuroinflammation, which may be heightened in ASD. Together, this suggests that the HS may also be altered in ASD. Using RNA sequencing (RNA-seq), we investigated genome-wide expression, as well as a focused gene set analysis of key HS genes (HDC, HNMT, HRH1, HRH2, HRH3 and HRH4) in postmortem dorsolateral prefrontal cortex (DLPFC) initially in 13 subjects with ASD and 39 matched controls. At the genome level, eight transcripts were differentially expressed (false discovery rate <0.05), six of which were small nucleolar RNAs (snoRNAs). There was no significant diagnosis effect on any of the individual HS genes but expression of the gene set of HNMT, HRH1, HRH2 and HRH3 was significantly altered. Curated HS gene sets were also significantly differentially expressed. Differential expression analysis of these gene sets in an independent RNA-seq ASD data set from DLPFC of 47 additional subjects confirmed these findings. Understanding the physiological relevance of an altered HS may suggest new therapeutic options for the treatment of ASD.
Similar content being viewed by others
Introduction
Autism spectrum disorder (ASD) is a class of neurodevelopmental disorders (NDDs) characterized by social impairment, repetitive behavior and impaired communication.1 Genetic studies indicate that the heritability of ASD is approximately 50–60%.2, 3 The underlying genetic architecture is complex, with risk conferred by many genetic and environmental factors. Many of these factors appear to be shared by other NDDs,4 consistent with a high level of comorbidity between ASD and other NDDs.5 Tourette syndrome (TS) shares genetic risk factors with ASD6, 7 and is among the most prevalently comorbid NDD with ASD.5 This may be, in part, due to shared upregulation in neuroinflammation,8, 9, 10 increased microglia activation11 and/or abnormalities in language processing related to frontal/basal ganglia circuit dysfunction.12, 13, 14 Indeed, boys with ASD show faster response times in procedural/grammar memory tasks similar to children with Tourette syndrome, which has been attributed to a reduction in the connectivity between the temporal lobe and the frontal/basal ganglia circuitry.12 Interestingly, evidence suggests that overactivation of microglia may result in the reduced connectivity seen in ASD.15
Each of these shared abnormalities may be mediated in part by the histaminergic system (HS). Histamine has a prominent role in nueroinflammation10, 16 and microglial regulation,11, 17, 18, 19 and recent studies suggest a role for the HS in TS, possibly reflecting alterations within basal ganglia circuitry.20, 21, 22 Critically, a single family study of TS demonstrated a causative role of a rare nonsense mutation in the histidine decarboxylase gene (HDC), which encodes the rate-limiting enzyme that converts histidine into histamine.20 Interestingly one of the TS subjects in this study also had a diagnosis of Asperger’s syndrome. Subsequent reports showed that two SNPs in the HDC region also associated with TS in a family genetic association study.23 Furthermore, recent studies of ASD also implicate potential involvement of HS in this disorder, as de novo deletions overlap histamine N-methyltransferase (HNMT), the gene encoding the enzyme that inactivates histamine.24, 25
The role of the HS in neuroinflammation, cognition, sleep, attention, sensory function and motor function has received considerable attention26, 27, 28 and recent evidence suggests that the HS may have a role in regulating microglia activation,11, 17 cytokine release18 and migration.19 Furthermore, the HS may influence microglia mediated phagocytosis and reactive oxygen species production through the activation of histamine receptors.29 Within the central nervous system (CNS), this system consists of histaminergic neurons within the tuberomammillary nucleus (TMN) of the hypothalamus that have diffuse projections into the spinal cord, brain stem and many telencephalic brain regions. Histamine synthesis occurs predominantly in the TMN, where HDC is largely expressed. In addition, mast cells and microglia also express HDC and contribute to brain histamine, particularly in response to inflammation.30, 31 Histamine inactivation by HNMT occurs throughout the brain. Four histamine G-protein-coupled receptors (H1R–H4R), encoded by the genes Histamine Receptor H1 (HRH1), Histamine Receptor H2 (HRH2), Histamine Receptor H3 (HRH3) and Histamine Receptor H4 (HRH4) are expressed throughout the CNS, although H4R expression is extremely low and its function within the CNS is not well characterized. Microglia express all four receptors and evidence suggests that histamine dose-dependently activates microglial activation and production of proinflammatory factors by binding H4R11 and possibly H1R.17
Antagonists/inverse agonists of H3R have been investigated as a therapy for a number of CNS diseases, including Alzheimer’s disease, epilepsy, narcolepsy, attention deficit hyperactivity disorder and fetal alcohol exposure-induced learning deficits.26, 32, 33, 34, 35 Unfortunately, clinical trial results have been largely disappointing; however, preclinical studies demonstrate promising results and early clinical trials of pitolisant showed encouraging results in the treatment of narcolepsy.34 Pitolisant may also prove efficacious in epilepsy, a condition highly comorbid with ASD.33 In addition, clinical studies demonstrate behavioral and sleep disturbance improvement in children and adolescents with ASD treated with H2R and H1R antagonists.36, 37
These therapeutic findings, combined with the recent genetic findings implicating HNMT in ASD and HDC in TS, the comorbidity of ASD and TS, and the overlap of a potential role for neuroinflammation, altered microglial activity and altered language processing between the two disorders raise the possibility that the HS is involved in ASD. We hypothesized, therefore, that the expression of histaminergic genes is altered in the brain in ASD. We focused on the expression of the following histaminergic genes: HDC, HNMT, HRH1, HRH2, HRH3 and HRH4 in the brains of subjects diagnosed with an ASD. Given the possibility of shared alterations in connectivity between the basal ganglia and frontal lobe in both TS and ASD, as well as the associated alterations in frontal lobe processing in general in ASD,29 we selected tissue samples from the dorsolateral prefrontal cortex (DLPFC). Based on the positive treatment findings in patients and animal models of ASD using H1R,37 H2R,36 and H3R35 antagonists, we predicted that the genes encoding these receptors would be more highly expressed in ASD subjects. We predicted that expression of HNMT would be altered, but we were unsure of the direction of change, given the overlap of HNMT with associated ASD deletions, and findings suggesting that polymorphisms of increased activity of HNMT are associated with modulating attention and hyperactivity behavior.38, 39
Materials and methods
A flow chart of the methods are outlined in Supplementary Figure 1.
Subjects and demographics
We evaluated the expression of histaminergic genes in the DLPFC of 13 individuals diagnosed with ASD and 39 non-psychiatric control subjects using deep RNA-seq coverage. Three control subjects were matched to each case subject by age (±6 years), gender and ethnicity when possible. Only five control subjects mismatched for ethnicity. A summary of subject demographic information is presented in Table 1 and more extensively in Supplementary Table 1.
The majority of samples were collected at the National Institute of Mental Health in the Section on Neuropathology, according to NIH protocol 90-M-0142, with informed consent of next-of-kin. Additional samples were consented through the University of Maryland Brain and Tissue Bank (formerly the NICHD Brain Bank), and one sample was consented through the Stanley Medical Research Institute. All the materials were transferred to the Lieber Institute for Brain Development under approved material transfer agreements. For details on National Institute of Mental Health brain collection, clinical characterization, neuropathological examinations and toxicological analyses, see Lipska et al.40 For details on the University of Maryland Brain and Tissue Bank and the Stanley Medical Research Institute sample characterization, see their respective websites at: www.medschool.umaryland.edu/btbank/ and www.stanleyresearch.org. Subjects with evidence of drug use, alcohol abuse or psychiatric illness were excluded from the control cohort. All the analyzed samples had RNA integrity number (RIN) values >6.
RNA extraction and sequencing
All RNA extraction and sequencing was performed at the Lieber Institute for Brain Development. RNA was extracted from postmortem homogenate brain tissue of gray matter dissected from Brodmann area 46 and 9. Total RNA was extracted using the RNeasy kit (Qiagen, Germantown, MD, USA) from approximately 100 mg of homogenate tissue. RNA-seq was performed using the TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero Gold ribosomal RNA depletion from Illumina (San Diego, CA, USA). One hundred base pair paired-end sequencing was run on the HiSeq 2000. The Illumina Real Time Analysis (RTA) module was used to perform image analysis and base calling and the BCL Converter (CASAVA v1.8.2) was used to generate the sequence reads. Sequencing depth was 40–60 million paired-end reads, yielding 80–120 million reads per sample. TopHat41 (v2.0.4) was used to enforce strand specificity and to align the sequencing reads to known transcripts of the Ensembl Build GRCh37.67, (a total of 57 659 genes with 196 495 known transcripts). Visual inspection of reads at each gene of interest was evaluated for each subject and no structural variants were found.
Differential expression analysis
We used featureCounts42 (v1.4.4) to count the total number of reads overlapping each gene using the default settings, with paired-end and reverse-stranded counting specified using the Ensembl Build GRCh37.67 gtf file. These counts were merged from both reads of the paired-end sequencing and normalized by library size and coding gene length to form Reads Per Kilobase of Gene per Million mapped reads (RPKM) values. These RPKM values were transformed using log2 after applying an offset of 1 to each count to stabilize the variance among lowly expressed genes and to prevent negative normalized counts, resulting in normalized gene expression levels: log2(RPKM+1). These normalized expression values were then filtered for genes with mean normalized expression values less than 0.5, reducing the total number of genes to 13 011. The influence of diagnosis status on all normalized and filtered gene abundance estimates was evaluated using a linear regression analysis covarying for principal components determined by the transcriptome-wide RNA-seq expression values for the 13 011 expressed Ensembl genes to control for potential technical artifacts and latent biological effects.
The appropriate number of principal components (PCs) to include in the model to correct for known and unknown confounds, yet still maintain variance related to diagnosis, was determined by the num.sv() function43 of the Bioconductor package ‘sva’ in R.44 This method uses parallel analysis to calculate the number of components that significantly contribute to overall variance, based on how often the eigenvalue for each PC is larger than eigenvalues calculated for random permutations of the data. This process is performed after removing variation due to specified variables of interest, in our case: diagnosis, RIN, sex, age, race and exonic mapping rate. This function determined that nine PCs significantly contributed to the overall variance. The percent variance contributed by each PC was as follows: 58.5, 9.2, 7, 3.8, 2.7, 2.3, 1.8, 1.5 and 1.2%, accounting for 88% of the overall variance. See Supplementary Figure 2 for a plot of the first two PCs and Supplementary Figure 3 for a heatmap indicating the association of each component with various known confounders.
After the model optimization, linear regression analysis was performed on the normalized gene expression levels across the transcriptome, modeling the effect of diagnosis while adjusting for the first nine PCs, as well as for age, RIN, sex, race and exonic mapping rate. Resulting t-statistics and corresponding P-values were calculated using the empirical Bayes method in the Bioconductor package ‘limma’ in R.45, 46 Multiple testing correction was performed using the Benjamini–Hochberg false discovery rate method. The histamine genes (HRH1, HRH2, HRH3, HRH4, HDC and HNMT) were then evaluated in the context of this genome-wide differential expression analysis. We also considered only male subjects (10 ASD and 30 controls) as sensitivity analyses to determine how much heterogeneity across the genders of the cases and controls mattered in the differential expression analysis, as recent studies indicate the potential existence of gender-specific protective and risk genetic factors.47 In the male subjects, we found that adjusting for the first seven PCs was the optimal number to include in the differential expression model to control for latent heterogeneity.
Gene set enrichment analysis
Given our interest in the impact of the HS as a pathway in ASD, we also evaluated these genes using gene set enrichment tests (GSETs). In addition, we considered five other curated histamine-related gene sets not based on our a priori hypotheses. HDC and HRH4 were more lowly expressed than the expression filter (<0.5) and were therefore not included in this analysis. We analyzed the expression of our target gene set including HRH1, HRH2, HRH3 and HNMT using the Bioconductor limma ROAST test, a rotation gene set test that is a self-contained gene set test and therefore tests whether the genes within the gene set are differentially expressed.48 The advantage of this test is that it does not rely on the assumption of gene independence48 and it has been found to be robust to small sample sizes and sample heterogeneity.49 Furthermore, this test is robust to small gene set sizes, as it does not rely on the hypergometric test, which tests for enrichment of the testing pre-specified gene set against all genes identified to have statistics below a significance threshold. Many gene set tests implement the hypergometric test, which can lead to inflated significance of small gene sets when one gene is highly differentially expressed. However, the ROAST test instead combines all single gene tests statistics into one set-level statistic, which is compared with calculated rotated gene set statistics. This test was performed using the normalized expression values and design matrix as used in the previous analyses. This test estimates significance based on random rotations of the orthogonalized residuals and provides output for three directional hypotheses: up, down and mixed. A mixed hypothesis indicates that the gene set includes extreme t-statistics and thus that genes within the gene set represent a large increase or decrease in expression.48 The other gene sets evaluated included three from gene ontology according to http://amigo.geneontology.org/: histamine receptor activity (GO:0004969), histamine secretion (GO:0001821) and histamine production involved in inflammatory response (GO:0002349), as well as two from GeneRIF identified using Harminizome:50 histamine and histaminergic. Genes within these gene sets that were expressed above the minimum expression threshold were evaluated in the GSETs. For a list of these genes, see Supplementary Table 2. We hypothesized that all gene set results would either be in the up or mixed direction, based on the positive treatment findings in patients and animal models of ASD using H1R,37 H2R36 and H3R35 antagonists and the inclusion of these genes or related genes in each gene set. In this analysis, 10 000 random rotations were run to obtain more precise statistical results. Significance values for the mixed and up hypothesis tests were corrected for multiple testing correction using the Benjamini–Hochberg false discovery rate method. The effect sizes of the q-values were obtained using the pes() function of the compute.es package51 in R.
Replication analysis
We performed a replication analysis using data recently published with independent samples in another recent ASD RNA-seq study.52 We received raw data from the Geschwind lab at UCLA for Ribo-Zero RNA-seq samples from 57 subjects including controls and cases with ASD and RIN >6 from the DLPFC, (Brodmann area 9). We excluded six ASD samples that were identified to have a 15q11.2-13.1 duplication, as the manifestation of ASD in these subjects may be due to a different pathogenesis. In addition, we excluded four samples because they overlapped with our original data set. Therefore, we used the raw data for a total of 47 subjects (28 controls and 19 patients). See Supplementary Table 3 for more extensive demographic data for these subjects.
Using these subjects allowed for a strict replication analysis, as the differential expression and GSETs were performed identically as in our data. The only difference was that these data were not strand-specific, and thus the setting used for counting was not strand-specific. Again, the num.sv() function43 of the Bioconductor package ‘sva’ in R was used to determine the optimal number of PCs to include in the model. This process was performed after removing variation due to specified variables of interest, in our case: diagnosis, RIN, sex, age and the percentage of assigned reads to Ensembl genes. This function determined that six PCs significantly contributed to the overall variance. The percent variance contributed by each PC was as follows: 32.9, 14.8, 12.8, 6.3, 5.2 and 3.8%, accounting for 75.8% of the overall variance. See Supplementary Figure 4 for a plot of the first two PCs and Supplementary Figure 5 for a heatmap indicating the association of each component with various known confounders. Only genes or gene sets with significant findings in the original data set were evaluated in the replication data. TRPC3 was not expressed in the replication data set and thus not tested when testing the GeneRIF histamine gene set.
Meta-analysis
Meta-analysis was performed using the Fisher’s method and the P-value statistics from both the original and the replication data sets using the sumlog() function of the ‘matap’ package53 in R. The histamine genes of interest, the genes that showed differential expression in the discovery analysis and the gene sets of interest that showed positive results in the discovery analysis were tested. Multiple testing correction was done using the Benjamini–Hochberg method. Importantly, this evaluation provides a measure for the statistical significance of the absolute value of gene expression changes across both the data sets but does not account for the directionality of the changes in expression.
Code availability
The code used to generate the results of this study is accessible at https://github.com/LieberInstitute/Wright_2017_ASD_Histamine. The information about data access can also be found at this site.
Results
Differential expression analyses reveal no difference in the expression of individual histamine genes
We identified 1463 genes differentially expressed between ASD subjects and healthy controls across all moderately expressed Ensembl genes (13 011) at marginal statistical (P<0.05) significance. Three control subjects were matched to each ASD subject for age and sex and nearly all subjects were also matched for ethnicity when possible. See Supplementary Table 4 for a full list of the test statistics for all evaluated genes. Following multiple testing correction, eight remained significantly differentially expressed at false discovery rate <5%: SNORA74A, SNORA53, SNORD17, TUBE1, SNORA54, SNORA74B, a lncRNA HSA-LNCG003387, also called RP6-206I17.3 or BX284650.2, and SNORD114-23, (Supplementary Table 5). None of the histamine genes of interest were significantly differentially expressed between cases and controls (Table 2); however, HNMT was nominally significantly differentially expressed, showing increased expression in ASD (P=0.003). HDC and HRH4 were excluded from the analysis as their mean estimated expression value was below the lower limit threshold (<0.5), as expected from prior knowledge of their expression. The analysis of only male subjects yielded 1027 differentially expressed genes before multiple testing correction, and three survived multiple testing correction including: SNORA54, SNORA74A and SNORA53 (false discovery rate <5%, Supplementary Table 6). The histaminergic gene results are presented in Supplementary Table 7.
We performed differential expression analysis using the same methods with the replication sample of 47 subjects. This yielded 1090 significantly differentially expressed genes before multiple testing correction (out of 13 473 that passed the minimum expression filter), and zero after multiple testing correction. Again, none of the histamine genes of interest were significantly differentially expressed individually following multiple testing correction, (Supplementary Table 8); however, HRH3 (P=0.013) was nominally significantly differentially expressed. HRH1 and HRH2 were found to be more lowly expressed in the patient group of the replication sample, and therefore showed the opposite direction of change as that of the original sample. None of the eight genes identified as significantly differentially expressed in our data were significantly differentially expressed in the replication data set at even nominal significance and roughly only half of these genes showed the same direction of fold change, as would be expected by chance, (Supplementary Table 9). The lack of consistency between the two data sets may be due to heterogeneity among samples and the relatively small size of our samples. Supplementary Table 10 shows the meta-analysis results for each of the histamine genes of interest and the genes identified to be significantly differentially expressed in the original data set. All differentially expressed genes identified in the original data set were also significant in the meta-analysis. HNMT (q=0.012), HRH1(q=0.033) and HRH3 (q=0.019) were also significant. However, HRH1 showed the opposite change of direction between the two data sets. HRH2 (q=0.296) was not significant in the meta-analysis.
Gene set enrichment discovery and replication analysis reveals that collectively the histamine genes show altered expression in the ASD subjects
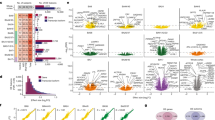
In contrast to the individual gene differential expression analyses, our target GSET of HNMT, HRH1, HRH2 and HRH3 using the ROAST test revealed that this gene set is significantly overexpressed (q-value =0.01), and significantly differentially expressed regardless of directionality (‘mixed’ test, q=0.02) in ASD patients, (Table 3). Evaluation of the fold changes within just the male subjects and our full data set in a sensitivity analysis indicates that the full data set was not biased by potential gender differences between diagnosis groups, (Supplementary Figure 6). Given the strength of the effect sizes, our results provide compelling evidence that the expression of the evaluated HS-related genes may be collectively altered in the DLPFC of ASD subjects. Furthermore, the replication analysis using the sample of 47 subjects confirmed that this gene set is similarly altered in expression in ASD prefrontal cortex. The gene set was found to be significantly different regardless of directionality also in this independent sample of ASD subjects, with the ROAST ‘mixed’ test (q-value =0.03, Table 4).
The Gene Ontology (GO) receptor gene set (GO:0004969) also was significantly upregulated (q=0.04), and both the GeneRIF gene sets showed significantly mixed expression (q=0.03 and q=0.02; Table 3) in the Lieber Institute for Brain Development sample. However, the GO histamine secretion (GO:0001821) and histamine production in inflammation (GO:0002349) gene sets did not show differential expression between the two subject groups. The GO receptor gene set and the GeneRIF histaminergic gene set also showed significantly mixed expression, (q=0.03 and q=0.03, Table 4) in the independent replication sample. See Figure 1 for a graphical representation of our target GSET in both the data sets. See Supplementary Figure 8 for a plot of the difference of the absolute log2 fold changes of these genes between the two data sets. Supplementary Table 10 shows the results of meta-analysis for the gene sets statistics across the two data sets. Our hypothesis-driven gene set using the ROAST up (q=0.004) and mixed (q=0.001) statistics were significant in the meta-analysis. In addition, the curated gene sets (evaluated in both data sets) using the ROAST mixed statistics were also significant.
Magnitude of altered expression of histamine-related genes in autism spectrum disorder (ASD). (a) The absolute log2 fold changes of gene expression between ASD subjects and control subjects within the original data set are shown on the x axis for all Ensembl genes evaluated in this differential expression analysis, while the y axis shows the frequency, or number of genes with each log2 fold change value. The x axis shown is limited to changes between 0 to 0.2 to allow easier evaluation of the genes of interest, however, the full range of absolute log2 fold changes was 4.9 × 10−6 to 1.2. This indicates the extent to which genes showed differential expression, regardless of the direction of change. The red lines indicate the absolute log2 fold changes of the histamine-related genes of interest. (b) The absolute log2 fold changes of gene expression between the ASD subjects and the control subjects within the replication data set are shown on the x axis for all Ensembl genes evaluated in this differential expression analysis, while the y axis shows the frequency, or number of genes with each log2 fold change value. The x axis shown is limited to changes between 0 to 0.2 to allow easier evaluation of the genes of interest, however, the full range of absolute log2 fold changes was 8.7 × 10−7 to 6.02. Again, the red lines indicate the absolute log2 fold changes identified for the genes of interest.
Discussion
Given recent HS findings in TS, the shared comorbidity between TS and ASD, the role of the HS in neuroinflammation and preliminary clinical trial data of histamine receptor inverse agonists/antagonists in ASD, we hypothesized that gene expression of histaminergic genes may be altered in postmortem brains of ASD patients. We evaluated the expression of individual HS genes as well as gene sets comprised of the genes encoding the more highly expressed histamine receptors in the CNS and the gene encoding the enzyme that inactivates and controls the activity of histamine.
None of our genes of interest were individually identified to be differentially expressed in the DLPFC of our sample or in our replication data set. Given the sample sizes involved, this is not inconsistent with what might have been expected. However, collectively as a gene set, our results suggest that the expression of these genes as representing a histaminergic signaling system, do show differential expression in the DLPFC of subjects with ASD compared with controls. Although this gene set had significant up and mixed ROAST results in the discovery sample, only the mixed result (of overall differential expression regardless of directionality) was replicated in the replication sample, an independent cohort of 47 subjects with RNA-seq data derived from Brodmann area 9. The directionality of a subset of specific genes varied across the discovery and replication data sets, yet the significance of overall differential expression suggests that the HS may be perturbed in ASD at a system level. Given that these genes regulate the response to histamine and therefore the influence of this neurotransmitter system on other neurotransmitters, such changes could be consequential for brain function and behavior. In addition, we identified several other histamine-related curated gene sets that showed significant differential expression regardless of directionality. Of note, we did not find significant differential expression of gene sets relating to histamine secretion or histamine production in response to inflammation. This suggests that perhaps the response to histamine is influenced in ASD, rather than its secretion or production in the brain due to inflammation. However, the GO production of histamine in response to inflammation gene set does not include HDC. Furthermore, our analyses did not evaluate the expression of this gene, as it was too lowly expressed. Therefore, production of histamine may still be altered, but this may not be due to a change of the tested inflammatory genes within this gene set. The HS system may still influence neuroinflammation rather than the other way around. Further research with additional and larger studies are necessary to characterize HS gene expression in ASD and to determine the potential consequence of altered gene expression of this signaling system. In addition, more research is required to characterize the HS and its interplay with neuroinflammation in ASD. Although the role of the HS is well described in systemic inflammation, the interaction of the HS with neuroinflammation in particular, is still being characterized. Evidence suggests that HRH1 may particularly be involved, however, the interplay may be highly contextually dependent.29
At the transcriptome level, our analysis yielded eight significantly differentially expressed individual genes, a number lower than one previous report54 yet comparable to another.55 These variable results are likely due to differences in the stringency of confounder correction. Our conservative method may obscure some true findings, yet as demonstrated in Supplementary Figure 7, the potential role of population confounders (for example, race) and of implicit RNA quality differences is not fully addressed by adjusting for typical demographic or RNA-seq quality control measures. Given our goal of testing differential expression of specific genes of interest, we opted for a conservative approach to minimize Type I error. Less stringent confounder correction may be more appropriate when testing more global differential expression hypotheses. Further work is needed to resolve the trade-offs in approaches to control for confounders in RNA-seq.
The top differentially expressed genes included small nucleolar RNAs (snoRNAs), which are interesting given recent findings of differentially methylated regions in paternal sperm and postmortem ASD human brain mapping to genes in the SNORD family.56 This warrants further investigation, but is beyond the scope of the current work. Interestingly, a deletion syndrome that included TUBE1 was associated with developmental delay and intellectual disability, suggesting the importance of this gene in brain development.57 To our knowledge, associations of SNORA74A, SNORA53, SNORD17, SNORA54, SNORA74B, RP6-206I17.3 and SNORD114-23 have not been previously reported in ASD.
There are important limitations of our study beyond the sample size, which though small in contrast to genetic population studies, is comparable to prior brain tissue studies of autism. First, many factors can influence the quantification of gene expression in postmortem tissue, including cellular composition, RNA quality, sequencing depth and so on. Evaluating cell-type-specific methylation signatures, using the methods as in Jaffe et al.58 indicated that there was no significant difference in the neuronal and microglial composition between our sample groups (data not shown), which is in agreement with results of another ASD DNA methylation study.59 In addition, the inclusion of transcriptome-wide principal components in our analyses reduces the influence of these known confounders as well as other unknown confounders. Furthermore, ASD is a very heterogeneous disorder, with potentially varied causative factors among different subjects. To consider this, we evaluated the copy number variations in a subset of our samples and identified that only 1 out of 10 evaluated ASD samples had a large copy number variation, which was located in chromosome 1. Importantly, none of the evaluated histamine genes of interest are located on this chromosome. We also excluded samples with the 15q11.2-13.1 duplication syndrome in our replication analysis. Our use of a gene set test that is robust to small sample size and sample heterogeniety49 should help mitigate some of the bias that may be introduced by each of these confounding effects. It would be especially important for future studies to investigate the expression of these histamine genes in neurons and microglia separately, to determine whether these genes are differentially expressed in primarily neurons, or microglia, or both. Deeper evaluation of the expression of each of the transcripts for these genes would also be helpful, as the shorter transcript of the HRH3, acts as an autoreceptor and therefore functions very differently than the longer isoforms. Deeper evaluation of the exon expression of HRH3 in our data indicate that only the longest transcripts were expressed. In the replication sample, the short autoreceptor transcript may have been expressed, although this is difficult to determine because it is unclear which exons are uniquely specific to the short form of the receptor. However, importantly all exons were more abundantly expressed in the ASD samples. Furthermore, expression studies cannot delineate between expression alterations due to causation or epiphenomena associated with the illness state of the condition studied.
HDC and HRH4 were not found to be highly expressed in the cortical regions evaluated in this study. Although this was expected, it would be interesting to evaluate the expression of these genes elsewhere. This is especially relevant for HDC, to determine whether it is under-expressed in the TMN, as it appears to be in particular cases of TS.7, 20, 21, 60 This possibility of altered receptor expression due to altered HDC expression is consistent with unpublished findings indicating that expression of HRH3 is elevated in the basal ganglia in Hdc knockout mouse models of TS.60 Another study of Hdc knockout mice found an increase in expression of HRH1 and HRH2 that was not significant, a significant decrease in expression of HRH3 in the hippocampus, and a significant increase in HRH3 expression in the TMN.61 Also, a human transcriptome study of the striatum found an increase in HRH3 that was not significant in TS subjects.62 Therefore, reduced HDC may induce regionally specific compensatory responses, although this requires further study. To our knowledge, the expression of HDC in the TMN has not yet been explored in ASD and it remains to be determined whether the expression of this gene is altered. Interestingly, higher plasma levels of histidine, the precursor of histamine, was identified in several ASD studies,63, 64 suggestive of peripheral alterations of histidine catabolism and histamine production. Histidine readily passes the blood–brain barrier; therefore, an overabundance of histidine peripherally could lead to increased histamine production in the CNS by HDC. A high level of histamine in the brain is associated with neuroinflammation, which is also observed in ASD and is predicted to be involved in the pathogenesis of many neurological diseases, including ASD.10, 65 Indeed TS may also have an inflammatory basis, as a recent study suggests enterovirus infection is associated with an increased incidence of Tic disorders;66 and similar to ASD,67 the incidence of allergies and asthma are strongly correlated with increased risk for TS.68 Further research is required to determine what role the HS may or may not have in the enhanced neuroinflammation observed in each of these disorders. Indeed changes in either neuroinflammation or the HS may simply be reflective of changes in the other, and potentially due to compensatory effects, rather than indicative of a causative association.
In conclusion, the HS is involved in modulating cognition and behavior and has a role in neuroinflammation. Our study provides, to our knowledge, the first specific evaluation of the expression of a set of histaminergic genes representing histamine signaling in the brain of ASD subjects. Many questions remain about the involvement of this system in ASD and the potential overlap in pathogenesis of ASD with TS. However, our results, which are replicated in an independent ASD postmortem data set, provide a starting point for the investigation of this system in ASD.
References
Brentani H, Paula CS, de, Bordini D, Rolim D, Sato F, Portolese J et al. Autism spectrum disorders: an overview on diagnosis and treatment. Rev Bras Psiquiatr 2013; 35: S62–S72.
Yoo H . Genetics of autism spectrum disorder: current status and possible clinical applications. Exp Neurobiol 2015; 24: 257.
Buehler MR . A proposed mechanism for autism: an aberrant neuroimmune response manifested as a psychiatric disorder. Med Hypotheses 2011; 76: 863–870.
De Rubeis S, Buxbaum JD . Genetics and genomics of autism spectrum disorder: embracing complexity. Hum Mol Genet 2015; 24: R24–R31.
Billstedt E, Gillberg C . Autism and Asperger syndrome: coexistence with other clinical disorders. Acta Psychiatr Scand 2000; 102: 321–330.
Clarke RA, Lee S, Eapen V . Pathogenetic model for Tourette syndrome delineates overlap with related neurodevelopmental disorders including Autism. Transl Psychiatry 2012; 2: e158.
Fernandez TV, Sanders SJ, Yurkiewicz IR, Ercan-Sencicek AG, Kim Y-S, Fishman DO et al. Rare copy number variants in tourette syndrome disrupt genes in histaminergic pathways and overlap with autism. Biol Psychiatry 2012; 71: 392–402.
Muller N . Tourette’s syndrome: clinical features, pathophysiology, and therapeutic approaches. Dialogues Clin Neurosci 2007; 9: 161.
Kern JK, Geier DA, Sykes LK, Geier MR . Relevance of neuroinflammation and encephalitis in autism. Front Cell Neurosci 2016; 9: 519.
Theoharides TC, Stewart JM, Panagiotidou S, Melamed I . Mast cells, brain inflammation and autism. Eur J Pharmacol 2015; 778: 96–102.
Frick L, Rapanelli M, Abbasi E, Ohtsu H, Pittenger C . Histamine regulation of microglia: Gene-environment interaction in the regulation of central nervous system inflammation. Brain Behav Immun 2016; 57: 326–337.
Walenski M, Mostofsky SH, Ullman MT . Inflectional morphology in high-functioning autism: evidence for speeded grammatical processing. Res Autism Spectr Disord 2014; 8: 1607–1621.
Prat CS, Stocco A, Neuhaus E, Kleinhans NM . Basal ganglia impairments in autism spectrum disorder are related to abnormal signal gating to prefrontal cortex. Neuropsychologia 2016; 91: 268–281.
Walenski M, Mostofsky SH, Ullman MT . Speeded processing of grammar and tool knowledge in Tourette’s syndrome. Neuropsychologia 2007; 45: 2447–2460.
Rodriguez JI, Kern JK . Evidence of microglial activation in autism and its possible role in brain underconnectivity. Neuron Glia Biol 2011; 7: 205–213.
Jutel M, Blaser K, Akdis CA . Histamine in allergic inflammation and immune modulation. Int Arch Allergy Immunol 2005; 137: 82–92.
Dong H, Zhang W, Zeng X, Hu G, Zhang H, He S et al. Histamine induces upregulated expression of histamine receptors and increases release of inflammatory mediators from microglia. Mol Neurobiol 2014; 49: 1487–1500.
Rocha SM, Pires J, Esteves M, Graça B, Bernardino L . Histamine: a new immunomodulatory player in the neuron-glia crosstalk. Front Cell Neurosci 2014; 8: 120.
Ferreira R, Santos T, Gonçalves J, Baltazar G, Ferreira L, Agasse F et al. Histamine modulates microglia function. J Neuroinflammation 2012; 9: 1.
Ercan-Sencicek AG, Stillman AA, Ghosh AK, Bilguvar K, O’Roak BJ, Mason CE et al. L-histidine decarboxylase and Tourette’s syndrome. N Engl J Med 2010; 362: 1901–1908.
Castellan Baldan L, Williams KA, Gallezot J-D, Pogorelov V, Rapanelli M, Crowley M et al. Histidine decarboxylase deficiency causes Tourette syndrome: parallel findings in humans and mice. Neuron 2014; 81: 77–90.
Bolam JP, Ellender TJ . Histamine and the striatum. Neuropharmacology 2015; 106: 74–84.
Karagiannidis I, Dehning S, Sandor P, Tarnok Z, Rizzo R, Wolanczyk T et al. Support of the histaminergic hypothesis in Tourette Syndrome: association of the histamine decarboxylase gene in a large sample of families. J Med Genet 2013; 50: 760–764.
Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung R-H et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 2012; 21: 3513–3523.
Mulatinho MV, de Carvalho Serao CL, Scalco F, Hardekopf D, Pekova S, Mrasek K et al. Severe intellectual disability, omphalocele, hypospadia and high blood pressure associated to a deletion at 2q22. 1q22. 3: case report. Mol Cytogenet 2012; 5: 1.
Witkin J, Nelson D . Selective histamine H3 receptor antagonists for treatment of cognitive deficiencies and other disorders of the central nervous system. Pharmacol Ther 2004; 103: 1–20.
Shan L, Bao A-M, Swaab DF . The human histaminergic system in neuropsychiatric disorders. Trends Neurosci 2015; 38: 167–177.
Haas HL, Sergeeva OA, Selbach O . Histamine in the nervous system. Physiol Rev 2008; 88: 1183–1241.
Rocha SM, Saraiva T, Cristóvão AC, Ferreira R, Santos T, Esteves M et al. Histamine induces microglia activation and dopaminergic neuronal toxicity via H1 receptor activation. J Neuroinflammation 2016; 13: 137.
Katoh Y, Niimi M, Yamamoto Y, Kawamura T, Morimoto-Ishizuka T, Sawada M et al. Histamine production by cultured microglial cells of the mouse. Neurosci Lett 2001; 305: 181–184.
Schwartz J-C, Arrang J-M, Garbarg M, Pollard H, Ruat M . Histamine transmission in the mammalian brain. Physiol Rev 1991; 71: 1–51.
Savage DD, Rosenberg MJ, Wolff CR, Akers KG, El-Emawy A, Staples MC et al. Effects of a novel cognition-enhancing agent on fetal ethanol-induced learning deficits. Alcohol Clin Exp Res 2010; 34: 1793–1802.
Kasteleijn-Nolst Trenité D, Parain D, Genton P, Masnou P, Schwartz J-C, Hirsch E . Efficacy of the histamine 3 receptor (H3R) antagonist pitolisant (formerly known as tiprolisant; BF2.649) in epilepsy: dose-dependent effects in the human photosensitivity model. Epilepsy Behav 2013; 28: 66–70.
Baronio D, Gonchoroski T, Castro K, Zanatta G, Gottfried C, Riesgo R . Histaminergic system in brain disorders: lessons from the translational approach and future perspectives. Ann Gen Psychiatry 2014; 13: 1–10.
Baronio D, Castro K, Gonchoroski T, de Melo GM, Nunes GDF, Bambini-Junior V et al. Effects of an H3R antagonist on the animal model of autism induced by prenatal exposure to valproic acid. PLoS ONE 2015; 10: e0116363.
Linday L, Tsiouris J, Cohen I, Shindledecker R, DeCresce R . Famotidine treatment of children with autistic spectrum disorders: pilot research using single subject research design. J Neural Transm 2001; 108: 593–611.
Rossi PG, Posar A, Parmeggiani A . Niaprazine in the treatment of autistic disorder. J Child Neurol 1999; 14: 547–550.
Gonzalez-Gadea ML, Chennu S, Bekinschtein TA, Rattazzi A, Beraudi A, Trippichio P et al. Predictive coding in autism spectrum disorder and attention deficit hyperactivity disorder. J Neurophysiol 2015; 114: 2625–2636.
Stevenson J, Sonuga-Barke E, McCann D, Grimshaw K, Parker KM, Rose-Zerilli MJ et al. The role of histamine degradation gene polymorphisms in moderating the effects of food additives on children’s ADHD symptoms. Am J Psychiatry 2010; 167: 1108–1115.
Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol Psychiatry 2006; 60: 650–658.
Trapnell C, Pachter L, Salzberg SL . TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25 (9): 1105–1111.
Liao Y, Smyth GK, Shi W . featureCounts:an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30 (7): 923–930.
Buja A, Eyuboglu N . Remarks on parallel analysis. Multivar Behav Res 1992; 27: 509–540.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD . sva: Surrogate Variable Analysis. R Package Version 3.8.0 2013 https://bioconductor.org/packages/release/bioc/html/sva.html.
Smyth GK . Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: 1–25.
Smyth GK . Limma: linear models for microarray data. Gentleman R, Carey V, Dudoit S, Irizarray R, Huber W (Eds.), Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer: New York, 2005; 397–420.
Lai M-C, Lombardo MV, Auyeung B, Chakrabarti B, Baron-Cohen S . Sex/gender differences and autism: setting the scene for future research. J Am Acad Child Adolesc Psychiatry 2015; 54: 11–24.
Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK . ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 2010; 26: 2176–2182.
Rahmatallah Y, Emmert-Streib F, Glazko G . Gene set analysis approaches for RNA-seq data: performance evaluation and application guideline. Brief Bioinform 2016; 17: 393–407.
Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016; 2016: baw100.
Del Re AC . compute.es: Compute Effect Sizes. R Package 2013 http://cran.r-project.org/web/packages/compute.es.
Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016; 540: 423–427.
Dewey M . metap: meta-analysis of significance values. R Package Version 0.8. 2017 https://CRAN.R-project.org/package=metap.
Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 2014; 159: 1511–1523.
Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun 2014; 5: 5748.
Feinberg JI, Bakulski KM, Jaffe AE, Tryggvadottir R, Brown SC, Goldman LR et al. Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. Int J Epidemiol 2015; 44: 1199–1210.
Tassano E, Mirabelli-Badenier M, Veneselli E, Puliti A, Lerone M, Vaccari CM et al. Clinical and molecular characterization of a patient with interstitial 6q21q22.1 deletion. Mol Cytogenet 2015; 8: 31.
Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016; 19: 40–47.
Ladd-Acosta C, Hansen KD, Briem E, Fallin MD, Kaufmann WE, Feinberg AP . Common DNA methylation alterations in multiple brain regions in autism. Mol Psychiatry 2014; 19: 862–871.
Rapanelli M, Pittenger C . Histamine and histamine receptors in Tourette syndrome and other neuropsychiatric conditions. Neuropharmacology 2015; 106: 85–90.
Chepkova A, Yanovsky E, Parmentier R, Ohtsu H, Haas HL, Lin J-S et al. Histamine receptor expression, hippocampal plasticity and ammonia in histidine decarboxylase knockout mice. Cell Mol Neurobiol 2012; 32: 17–25.
Lennington JB, Coppola G, Kataoka-Sasaki Y, Fernandez TV, Palejev D, Li Y et al. Transcriptome analysis of the human striatum in Tourette syndrome. Biol Psychiatry 2016; 79: 372–382.
Kotsopoulos S, Kutty KM . Histidinemia and infantile autism. J Autism Dev Disord 9: 55–60.
Naushad SM, Jain JM, Prasad CK, Naik U, Akella RR . Autistic children exhibit distinct plasma amino acid profile. Indian J Biochem Biophys 2013; 50: 474–478.
Bañuelos-Cabrera I, Valle-Dorado MG, Aldana BI, Orozco-Suárez SA, Rocha L . Role of histaminergic system in blood–brain barrier dysfunction associated with neurological disorders. Arch Med Res 2014; 45: 677–686.
Tsai C-S, Yang Y-H, Huang K-Y, Lee Y, McIntyre RS, Chen VC-H . Association of Tic disorders and enterovirus infection: a nationwide population-based study. Medicine (Baltimore) 2016; 95: e3347.
Theoharides TC, Tsilioni I, Patel AB, Doyle R . Atopic diseases and inflammation of the brain in the pathogenesis of autism spectrum disorders. Transl Psychiatry 2016; 6: e844.
Chang Y-T, Li Y-F, Muo C-H, Chen S-C, Chin Z-N, Kuo H-T et al. Correlation of Tourette syndrome and allergic disease: nationwide population-based case-control study. J Dev Behav Pediatr 2011; 32: 98–102.
Acknowledgements
We greatly appreciate the Geschwind lab for providing us access to their original bam files for our replication analysis. We would like to gratefully acknowledge the families of the subjects whose donations made this research possible. We thank the National Institute of Mental Health, Division of Intramural Research, the University of Maryland Brain and Tissue Bank and the Stanley Medical Research Institute for their contributions to this research. We are grateful for the generosity of the Lieber and Maltz families in establishing an institute dedicated to understanding the basis of developmental brain disorders. This research was funded by the Lieber Institute for Brain Development and the AstraZeneca postdoctoral fellowship program. This research was also funded in part by the following grant: 2 T32 MH 15330-37 (PI: DRW).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
AJC and NJB are full-time employees and share holders of AstraZeneca PLC. CW is a postdoctoral fellow in the AstraZeneca postdoctoral studentship program. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Wright, C., Shin, J., Rajpurohit, A. et al. Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiatry 7, e1126 (2017). https://doi.org/10.1038/tp.2017.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2017.87