Abstract
Dopamine function is broadly implicated in multiple neuropsychiatric conditions believed to have a genetic basis. Although a few positron emission tomography (PET) studies have investigated the impact of single-nucleotide polymorphisms (SNPs) in the dopamine D2 receptor gene (DRD2) on D2/3 receptor availability (binding potential, BPND), these studies have often been limited by small sample size. Furthermore, the most commonly studied SNP in D2/3 BPND (Taq1A) is not located in the DRD2 gene itself, suggesting that its linkage with other DRD2 SNPs may explain previous PET findings. Here, in the largest PET genetic study to date (n=84), we tested for effects of the C957T and -141C Ins/Del SNPs (located within DRD2) as well as Taq1A on BPND of the high-affinity D2 receptor tracer 18F-Fallypride. In a whole-brain voxelwise analysis, we found a positive linear effect of C957T T allele status on striatal BPND bilaterally. The multilocus genetic scores containing C957T and one or both of the other SNPs produced qualitatively similar striatal results to C957T alone. The number of C957T T alleles predicted BPND in anatomically defined putamen and ventral striatum (but not caudate) regions of interest, suggesting some regional specificity of effects in the striatum. By contrast, no significant effects arose in cortical regions. Taken together, our data support the critical role of C957T in striatal D2/3 receptor availability. This work has implications for a number of psychiatric conditions in which dopamine signaling and variation in C957T status have been implicated, including schizophrenia and substance use disorders.
Similar content being viewed by others
Introduction
Genetic variation in the dopamine (DA) D2 receptor (DRD2) gene or its neighbor, the ankyrin repeat and kinase domain containing 1 (ANKK1) gene, have been associated with risk for schizophrenia1, 2, 3, 4 and its response to pharmacological treatment.5, 6, 7, 8, 9 As most antipsychotics used to treat the positive symptoms of schizophrenia act on D2 receptors,10, 11, 12, 13, 14, 15, 16, 17 a better understanding of how genetic variation may affect D2/3 receptor availability (binding potential, BPND, a ratio of specifically bound D2/3 tracer to its nondisplaceable concentration) could be useful in aiding clinicians in prescribing more targeted treatments. The C957T (rs6277) single-nucleotide polymorphism (SNP) of the DRD2 gene warrants particular attention given that the C allele has been associated with schizophrenia in Caucasians in a meta-analysis of nearly 7000 participants (3000 schizophrenia cases).18 Understanding the functional consequences of the C957T SNP has implications beyond schizophrenia as this SNP has also been associated with an increased risk for substance use disorders.19, 20, 21 However, to date only one research group has reported on the effect of this SNP on D2/3 BPND.22, 23, 24 Another DRD2 SNP, -141C Ins/Del (rs1799732), has also been reported to impact striatal D2/3 BPND with Jonsson et al.25 finding heightened BPND in Del carriers, but replication has been difficult, with Pohjalainen et al.26 finding no significant effect (and, if anything, higher BPND in Ins/Ins versus Ins/Del individuals) despite a similarly sized sample and use of the same radiotracer, 11C-raclopride. To date, more attention has been paid to the impact of the Taq1A SNP in the ANKK1 gene (rs1800497) with several groups reporting that A1 carriers have lowered striatal D2/3 BPND relative to A2 homozygotes.25, 27, 28 However, sample sizes in these studies have generally been small and the results have not been consistent across all studies.29, 30, 31 Nevertheless, based on the positive findings in the literature, many researchers have used Taq1A as a proxy for D2 receptor status (or more loosely as an index of general dopamine functioning32, 33, 34, 35).
Given that Taq1A polymorphism does not occur within the DRD2 gene itself, researchers have speculated that polymorphisms in Taq1A may associate with other SNPs in the DRD2 gene that are the real drivers of expression of the receptor in vivo.36 The C957T and -141C Ins/Del polymorphisms are in strong linkage disequilibrium with Taq1A21, 37, 38 and have themselves been associated with striatal D2/3 BPND.22, 23, 25 Despite the data suggesting that these SNPs are strongly linked, few studies have systematically investigated the effect of C957T, -141C Ins/Del, and Taq1A in isolation and combination on D2/3 BPND. Characterizing the functional effect of these SNPs on D2/3 BPND has implications for better understanding the mechanisms through which they exert their demonstrated influence on motivated behaviors including learning and decision making.39, 40, 41
Furthermore, as past research has implicated C957T,22, 23 Taq1A25, 27, 28, 42 and -141C Ins/Del25 in striatal BPND, the use of multilocus SNP scores that combine the effects of each DRD2 variant might be useful to understand whether these SNPs have additive effects on BPND. Multilocus dopaminergic scores have been used in a number of behavioral/clinical and functional magnetic resonance imaging studies,43, 44, 45, 46, 47, 48 but have surprisingly not been conducted in dopamine imaging. A multilocus approach provides an added advantage of determining the relative impact of each SNP on D2/3 BPND. Furthermore, given that the majority of the previous studies used the positron emission tomography (PET) radiotracer 11C-raclopride, which is not able to image extrastriatal BPND, little is known about the impact of these DRD2 SNPs on D2/3 receptor availability outside the striatum. Although one paper has investigated extrastriatal D2/3 BPND using 11C-FLB-457 and found an effect of C957T,24 it was limited by low numbers of CC (n=7) and TT (n=8) individuals in the analysis. Considering that there is evidence that striatal and extrastriatal D2/3 receptors are differentially regulated,49 further exploration of the effects of DRD2 SNPs on receptor availability across the brain is needed. In the present study, we used 18F-Fallypride, which is a D2/3 receptor tracer with favorable affinity to measure both striatal and extrastriatal receptors. We assessed the impact of C957T, Taq1A and -141C Ins/Del SNPs and multilocus effects of these SNPs in combination on D2/3 BPND in a sample of 84 healthy subjects.
Materials and methods
Subjects
Our data set consisted of 84 total participants (ages 18–37, m=24.17±5.05; 53.6% female; 69% Caucasian) who participated in three PET studies in the Zald Affective Neuroscience lab over the period of 10 years. Participants gave written informed consent, as approved by the Vanderbilt University Institutional Review Board.
Participants had no known past or present neurological or psychiatric diagnoses, no history of substance use disorders and no current use of psychoactive medications or substances as assessed by Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders I50 administered at screening.
PET imaging
[18F]-Fallypride ((S)-N-[(1-allyl-2-pyrrolidinyl)methyl]-5-(3[18F]fluoropropyl)-2,3-dimethoxybenzamide) was produced in the radiochemistry laboratory attached to the PET unit at Vanderbilt University Medical Center, following synthesis and quality control procedures described in the US Food and Drug Administration IND 47 245. All the data were collected on the same GE Discovery STE PET scanner.
Serial scan acquisition was started simultaneously with a 5.0 mCi (185 MBq) slow bolus injection of DA D2/3 tracer [18F]-Fallypride (specific activity >3000 Ci mmol−1). Computed tomographic scans were collected for attenuation correction before each of the three emission scans, which together lasted approximately 3.5 h with two breaks for subject comfort. Acquisition times for the dynamic PET scans were the same across all studies and have been reported previously.51
PET data processing
After decay correction and attenuation correction, PET scan frames were corrected for motion using SPM8 (ref. 52) with the last dynamic image frame of the first series serving as the reference image. The mean PET image created from the realignment was then registered to each subject’s high-resolution T1 magnetic resonance image (FLIRT, 6 degrees of freedom), which was nonlinearly registered to MNI space (FNIRT) in FSL.53 Putamen and cerebellum reference regions of interest (ROIs) were created from the WFU Pickatlas54 with the cerebellum modified such that the anterior one-fourth of the ROI along with voxels within 5 mm of cortex were excluded to prevent contamination of the PET signal from nearby areas such as midbrain or occipital cortex. These ROIs were then warped to each subject’s PET space using the FLIRT and FNIRT FSL transform matrices (MNI→T1→PET) and used in a simplified reference tissue model55 performed in PMOD software (PMOD Technologies, Zurich, Switzerland) to estimate Fallypride binding potential (BPND, a ratio of specifically bound Fallypride to its nondisplaceable concentration). Specifically PMOD’s PXMOD tool was used to estimate BPND voxelwise using a published basis function fitting approach.56
Subject-specific BPND images were then warped to MNI space using the saved FSL transforms to create MNI-normalized BPND images (resampled to 2 mm isotropic voxels). These MNI-normalized images were then analyzed (using an explicit MNI brain mask) in SPM8 to test for their relation to SNPs in the DRD2 gene.
Genotyping of DRD2 SNPs
Blood samples from each subject were genotyped for Taq1A (rs1800497), C957T (rs6277) and -141C Ins/Del (rs1799732) SNPs via Sequenom analysis performed at Vanderbilt University’s VANTAGE Genomics Core (see ref. 57 for detailed Sequenom genotyping methods).
PET analyses for DRD2 SNP effects
In all the analyses, we controlled for age and sex as these have been found to affect dopamine signaling.58, 59, 60, 61 We initially performed independent sample T-tests in SPM8 comparing BPND for Taq1A A2A2 versus A1 Carriers as well as -141C Ins/Ins vs Del Carriers as these groupings have often been used when analyzing these two SNPs.25, 27 We also tested for a linear effect of A2 allele dosage given previously published data.62 For the C957T SNP, we tested for linear effects of T allele dosage via a multiple regression analysis in SPM with number of T alleles as our independent variable of interest. We had a priori hypotheses that the three SNPs would affect striatal BPND given previously published 11C-raclopride PET data.22, 23, 25, 27 Therefore, we also applied a small volume correction in all SPM8 analyses that consisted of a bilateral striatal ROI composed of caudate, putamen and ventral striatum as defined in Mawlawi et al.63 and used in prior PET studies,64, 65, 66 thus limiting significance testing to the striatum by masking the SPM T images in follow-up analyses. We also explored the effects of additive multilocus scores comprising our DRD2 SNPs (weighted as in previously published PET studies or based on our own single SNP analyses when our data did not conform to previous reports, which was the case with the Ins/Del SNP) via multiple regression of allelic dose with Fallypride BPND. To clarify the results, we investigated BPND extracted from the anatomical striatal ROIs63 in post hoc analyses when significant effects were observed in the striatum during the primary voxelwise analyses. In supplemental analyses, we also extracted BPND from anatomical masks of extrastriatal regions (see Supplementary Information for details).67 We also calculated η2 effect sizes (controlling for age and sex) and 95% confidence intervals for BPND obtained from both our striatal and extrastriatal ROIs across genotype groups to allow for comparisons with previously published findings.42
Results
DRD2 SNP distributions and associations
All SNPs were present in expected ratios and did not violate Hardy–Weinberg equilibrium (max χ2=4.94, min P=0.09 for Ins/Del; see Table 1). There were significant differences in the Taq1A distribution across the C957T individuals (χ2=14.66, df=4, P=0.005) with A1A1 being exclusively present in CC individuals and the majority of TT individuals expressing A2A2 (79%, 11/14). There was a trend toward differences in Taq1A distributions across -141C Ins/Del group (χ2=8.02, P=0.091), but this was undoubtedly driven by the lack of individuals with two copies of either rare alleles (Del (~5%) and A1 (~7%)). When comparing distributions of Taq1A A1 Carriers vs A2A2, no difference in Ins/Del genotype distribution was present (χ2=0.31, df=2, P=0.86). There was, however, a significant difference in C957T distribution across Ins/Del individuals (χ2=12.77, df=4, P=0.012) with all TT individuals expressing Ins/Ins (14/14) and CC individuals being majority Del/Del (75%, 3/4).
Importantly, there were no significant differences in sex distributions or age across our genotype groups (Table 1), whereas differences in ethnicity across C957T and Taq1A were expected given previously reported allelic distributions by ethnic group.68 Covarying for participant ethnicity (Caucasian, African American, Asian, or Hispanic), however, did not alter the statistical significance or lack thereof of any reported results.
C957T and Fallypride BPND
Controlling for participant age and sex in our voxelwise analysis, we found two large striatal clusters in which BPND increased along with the number of C957T T alleles. The clusters, one in each hemisphere, both reached cluster-level false discovery rate (FDR) significance: (i) k(voxel #)=528, T=4.48, PFDR=0.018, peak at 26, 10, −4; and (ii) k=516, T=3.86, PFDR=0.018, peak at −24, 4, −2 (Figure 1). The left striatal cluster also extended down into the ventral striatum. These results were supported by anatomically based striatal ROI analysis, which showed that BPND differed significantly in the putamen and ventral striatum (VS, Supplementary Table S1, Supplementary Material). We found no support for C957T effects on extrastriatal BPND in our voxelwise analysis, except for a very small set of voxels in the midbrain/thalamus (k=39; Figure 1). Because Hirvonen et al.24 reported extrastriatal C957T effects using a priori cortical ROIs, which can be more sensitive to group effects due to their use of a more stable regional aggregate of BPND, we further tested for an effect of C957T on extrastriatal ROIs. However, we found no significant differences in BPND in these extrastriatal ROIs (Supplementary Table S2).
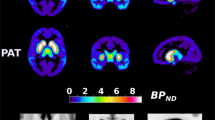
C957T T allele dosage is associated with increased striatal BPND. Results from a regression analysis run in SPM8 identified areas where Fallypride BPND was positively correlated with number of T alleles in the C957T SNP. Large clusters were observed in the striatum with both left and right clusters surviving an FDR cluster-level correction for multiple comparisons. A small (k=39) midbrain/thalamic cluster (peak at 2, −10, −2) is visible on the axial slice. In all figures, data are displayed in neurological convention (image on left represents left side of brain). Data are displayed using a P<0.005, uncorrected threshold. BPND, nondisplaceable binding potential; FDR, false discovery rate; SNP, single-nucleotide polymorphism.
Taq1A and Fallypride BPND
Investigating the effect of Taq1A on Fallypride BPND, an A2A2>A1 Carrier T-test run in SPM resulted in no significant clusters, even at cluster-level P<0.05 uncorrected. We also tested for a linear effect of A2 dose on BPND (A1A1<A1A2<A2A2) via regression in SPM. Again, no significant clusters were identified. In neither case did we identify significant effects of Taq1A on BPND after applying a small volume correction within our striatal ROI. We also found no evidence for genotype effects in BPND in ROI analysis of the three striatal subregions (Supplementary Table S3).
141C Ins/Del and Fallypride BPND
When comparing -141C Ins/Del Del Carriers with Ins/Ins, no significant clusters were present in the Del Carriers>Ins/Ins BPND analysis (where a previous effect had been observed on striatal BPND25). The opposite contrast, Ins/Ins>Del Carriers, resulted in two modest clusters, but neither were significant after correcting for multiple comparisons: right sub-gyral/orbitofrontal cortex (k=264, T=3.63 at 24, 28, −8, PFDR cluster level=0.32) and midbrain/pons (k=111, T=3.38 at 0, −26, −28, PFDR cluster level=0.82). We observed no significant BPND effects in the striatum even after applying a small volume correction. We also found no evidence for genotype differences in BPND across striatal ROIs (Supplementary Table S4).
DRD2 multilocus analyses: C957T alone explains BPND effect in striatum
Given that all of the DRD2 SNPs that we examined are believed to be in high linkage disequilibrium,21, 37, 38 we tested whether the addition of either Taq1A or -141C Ins/Del genotype, or both, increased the prediction of BPND beyond the observed effects of the C957T SNP. The addition of Taq1A genotype to C957T did not provide additional benefit in predicting Fallypride BPND (Supplementary Table S5). The addition of Ins/Del Ins/Ins vs Del Carrier status to C957T T allele dose (T allele #+Ins/Ins status (0,1)) increased the spatial extent of right striatal voxels in which BPND was associated with genotype (k went from 528 to 1019) but not the strength of the association (max T value went from 4.48 to 4.20 (PFDR=0.002 at 26, 8, −4); Supplementary Table S6). The lack of improvement in strength of association was confirmed by ROI analysis (see Supplementary Materials). The left striatum effect decreased in both spatial extent (k went from 516 to 488) and strength (max T value went from 3.86 to 3.41; PFDR 0.018 to 0.043 at −20, 8, −8). In addition, the combined C957T+Ins/Del score was associated with higher BPND in a large midbrain/pons area (k=353, T=4.30 (global max from this analysis) at 2, −26, −28), though it did not survive corrections for multiple comparisons (PFDR=0.087), and did not conform to the location and shape of a specific anatomical structure (Figure 2; Supplementary Table S6), although it is notable that part of the focus was in the vicinity of the raphe nuclei. Furthermore, a small midbrain/thalamus area was identified (k=144, T=3.70 at 8, −22, −4) but did not reach significance (PFDR=0.53; Figure 3, Supplementary Figure S1).
Multilocus DRD2 SNP effects on BPND. C957T T+Ins/Ins score (alone on left, displayed in hot yellow colors) and C957T T+Ins/Ins+Taq1A A2 dose score (center, displayed in cool colors) effects on Fallypride BPND are displayed as T-scores. Beyond the striatum, the C957T T+Ins/Ins multilocus score affects BPND in midbrain/thalamus and midbrain/pons (evident in coronal and saggital slices). The addition of A2 dosage information does not alter the multilocus score’s relationship to striatal and thalamic BPND appreciatively (overlap of both multilocus effects in white/pink). Taq1A A2 dose also has no effect on the deeper midbrain/pons genetic effect observed with the C957T+Ins/Ins multilocus score (see saggital slice). Data are displayed using a P<0.005, uncorrected threshold. BPND, nondisplaceable binding potential; SNP, single-nucleotide polymorphism.
Fallypride BPND increases with increasing T allele number in the ventral striatum and putamen. Graphs depict Fallypride BPND (covarying for age and sex) from anatomically based striatal ROIs as defined by Mawlawi et al.63 plotted by C957T genotype. Significant linear effects of T allele dosage were observed for right (β=0.32, P=0.004) and left (β=0.27, P=0.015) putamen as well as right (β=0.22, P=0.039) and left (β=0.35, P=0.001) ventral striatum. Note minimum BPND is set at 10 in all figures to better display group differences. Error bars reflect s.e.m. **P<0.01, *P<0.05, #P<0.10 reflect significance of across genotype group differences in BPND, controlling for age and sex. BPND, nondisplaceable binding potential; ROI, region of interest; SNP, single-nucleotide polymorphism; VS, ventral striatum.
Investigating the effect of a combined multilocus score with number of C957T T alleles, 141C Ins/Ins status (1,0) and number of Taq1A A2 alleles on Fallypride BPND resulted in qualitatively similar results in the striatum as our C957T analysis alone (Supplementary Table S7). Furthermore, stepwise regression, with age and sex in the first step, and C957T in the second step, revealed that there was no significant improvement in predictive power in the identified left (F-change=0.333, P=0.566) or right (F-change=0.775, P=0.381) striatum, above the effects of C957T, when Ins/Ins status or number of Taq1A A2 alleles were added in the third and fourth steps. We also conducted anatomically based striatal ROI stepwise regression analyses that confirmed that C957T explains more of the variance in BPND than the Taq1A and -141C Ins/Del SNPs, particularly in ventral striatum and putamen (see Supplementary Materials).
Discussion
C957T T allele is associated with heightened striatal D2/3 receptor availability
Here, we demonstrate that increasing number of C957T T alleles are associated with heightened D2/3 receptor availability (BPND) in large portions of the striatum. Our results replicate the previous observation with 11C-raclopride PET that C957T T allele dosage is related to higher BPND in the striatum.22, 23 Such replications are critical in PET studies because the expense and inconveniences of PET radioligand research leave most studies substantially underpowered for genetic analysis. However, the directness of the links between genes for a given receptor and PET assessment of those same receptors makes SNPs such as C957T (in the DRD2 gene itself) more reasonable targets for genomic neuroimaging than most candidate polymorphisms. It is notable that we observed the C957T effect with a different D2/3 radiotracer (18F-Fallypride) than Hirvonen et al.22, 23 (11C-raclopride), further suggesting the robustness of the effect. We also report for we believe the first time the effect of C957T in predicting D2/3 BPND in specific subregions of the striatum63, 64, 65, 66 and found support for the T allele being associated with higher bilateral putamen and ventral striatum BPND (but only restricted impact in the caudate).
C957T and extrastriatal D2/3 receptor availability
A primary advantage of [18F]-Fallypride over [11C]-raclopride as a tracer is its ability to measure extrastriatal D2 receptors. We therefore sought to replicate the findings of Hirvonen et al.24 who found that C alleles were associated with higher 11C-FLB-457 binding in anatomically defined extrastriatal regions. However, our voxelwise analysis did not identify any significant extrastriatal clusters, and we found no evidence for differences in BPND in extrastriatal ROIs chosen to approximate those of Hirvonen et al.24 Although qualitatively BPND in some cortical ROIs was higher with the C allele, as found by Hirvonen et al.,24 they did not reach statistical significance. Thus, C957T is not exerting a homogeneous global influence over both striatal and extrastriatal regions. This is consistent with evidence that individual differences in the striatal and cortical D2 BPND are at least partially dissociable,49 which in turn suggests that some genetic and environmental influences on D2 receptor expression and functioning should be expected to be different across regions.
Reconciling PET and in vitro data on C957T
One reason why our replication of the prior striatal findings of Hirvonen et al.22, 23 is important is that the direction of the C957T effect in the striatum is opposite of what would be predicted based on in vitro data where the T allele in the synonymous C957T SNP in CHO-K1 cells is associated with less DRD2 protein synthesis and less stable DRD2 mRNA (due to folding).37 The source of the discrepancy between the in vitro data and the striatal PET data is unclear. The CHO-K1 cell line used is nonhuman in origin (from hamsters), does not normally express DRD2, and may potentially be a poor proxy for human cells that naturally express D2 receptors in striatum (medium spiny neurons). Taken together, the human PET data strongly suggest that it is a mistake to extrapolate the in vitro finding of Duan et al.37 to human striatal D2 receptor expression in vivo.
Moderate effect of Ins/Del SNP on striatal and midbrain/pons D2/3 receptor availability
The potential role of DRD2 SNPs in affecting D2/3 BPND in extrastriatal subcortical regions will require further study, as our results are somewhat equivocal and did not reach conservative levels of statistical significance. Our voxelwise data suggest the -141C Ins/Del SNP may affect BPND (Ins/Ins>Del carriers) in the midbrain/pons even though it had little effect in the striatum (η2=0.007, d=0.17 from ROI analysis, Supplementary Table S4). Previous work has found only minor25 or no effect26 of Ins/Del genotype on striatal BPND. Specifically, Jonsson et al.25 observed a small (P=0.024; Cohen’s d~0.69) effect of -141C Ins/Del with Del Carriers having higher striatal D2/3 BPND, opposite to the effect we observe here. Their data, however, were collected across two different PET scanners, which could have introduced systematic variance in the data (see the ‘Lack of Robust Effect of Taq1A’ section below). A similarly sized raclopride PET study observed no significant effect of Ins/Del on striatal BPND but the direction of difference was similar to what we observed (higher for InsIns).26 One reason for this discrepancy may be that neither study reported data from the different striatal subdivisions. In contrast to the ventral striatum and putamen, we observed slightly higher BPND in the caudate of Del Carriers, suggesting that averaging across striatal subdivisions may mask the SNP’s effects. Finally, we note that our voxelwise results of increased D2/3 receptor availability (BPND) in Ins/Ins individuals fits with in vitro data using two human-derived cell lines, including Y-79 cells demonstrated to express functional D2 receptors,69 which show that the Del variant in -141C results in reduced transcriptional efficiency of the DRD2 gene.1
Lack of robust effect of Taq1A on D2/3 receptor availability
Although a Taq1A A2/A2>A1 Carriers effect on striatal BPND has been observed in a recent meta-analysis of five studies42 and our dataset had ~80% power to observe the mean effect size of d=0.57, we found no effect of Taq1A genotype on Fallypride BPND in our voxelwise analysis. Furthermore, our ROI analysis found only a very small A2/A2>A1 BPND effect (Hedges g=0.12, 95% confidence interval: −0.21, 0.28) in the striatum that was around 20% of that reported in the meta-analysis42 with the confidence interval including zero, suggesting that the effect was not robust. The authors of the meta-analysis point out that certain moderators, including age and sex, might explain variation in PET/SPECT studies focused on the relationship between DRD2 genetics and D2/3 BPND. Importantly, when we controlled for sex and age effects in our ROI analyses, we observed no effect of Taq1A on striatal BPND (min P=0.24; max η2=0.017, d≈0.26; Supplementary Table S3). Earlier studies observing Taq1A effects have often not controlled for these potential confounds on BPND. Furthermore, not all imaging studies have found effects of Taq1A on BPND including the study with the largest sample size to date (n=70)29, 30, 31 and at least one of the most-cited studies showing an effect has a methodological concern. That study, by Jonsson et al.,25 consisted of half the sample being run on a different PET system, which they tried to correct for with a systematic multiplication to their data (bound/free ratio). This approach could have introduced systematic error in the data as the paper does not provide the distribution of the genotypes across the two PET scanners used. Here, we limited our genetic analyses to data collected on the same PET system—a GE Discovery STE. The present study is also the largest (n=84) single PET study on DRD2 genetic effects to date. Our systematic analysis suggests that Taq1A allele status does not robustly affect D2/3 BPND except in specific striatal subdivisions and, thus, raises caution in the use of this SNP as a proxy for global striatal D2 receptor levels (or for DA functioning more generally) as has been the case in some of the literature.32, 33
Although this study utilized the D2/3 tracer 18F-Fallypride (vs 11C-raclopride in most prior studies), we do not have a reason to specifically predict that kinetic properties of the D2/3 tracer used would lead to a different result. That said, Fallypride has higher affinity for D2-like receptors and appears less sensitive to endogenous dopamine levels than raclopride.70, 71 Thus, if there are indeed significant effects of Taq1A on raclopride binding but not Fallypride binding, it could suggest that Taq1A effects are due to an impact on endogenous dopamine levels, rather than DRD2 affinity or receptor density. In fact, an 18F-DOPA PET study has implicated the A1 allele of Taq1A (but no effect of C957T or -141C Ins/Del SNPs) with increased dopamine synthesis in the putamen.72 Interestingly, a recent PET study using the D2-specific radiotracer 11C-NMB, which is relatively insensitive to endogenous DA levels, did observe an effect of Taq1A on striatal BPND.73 However interpretation of this study is complicated by the fact that only 24 of the 57 participants studied were considered healthy controls and disease state may influence Taq1A effects on BPND.42 That paper also did not examine effects for SNPs other than Taq1A, and thus did not address whether C957T status affects striatal BPND. Clearly, further work is needed to determine the biological processes underlying differences in BPND observed with various PET tracers as well as the role of specific DRD2 SNPs on these processes.
Linkage of DRD2 SNPs
In our data, individuals expressing the Taq1A A2 allele were more likely to also express the C957T T and Ins/Del Ins alleles. Others have reported strong linkage disequilibrium between C957T, -141C Ins/Del and Taq1A21, 37 or between C957T and Taq1A.38 To follow up on this work, we used LDmatrix74 to search the 1000 Genomes population database across all HapMap ethnic stratifications, and found linkage disequilibrium to be much higher between C957T and Taq1A (D′=0.76) and C957T and -141C Ins/Del (D′=0.84) than between Taq1A and -141C Ins/Del (D′=0.12). Thus, there is strong empirical evidence that C957T is linked with two other SNPs where D2/3 BPND effects have been observed with PET/SPECT42, 75 and, therefore, may have driven some of the effects observed with Taq1A (or -141C Ins/Del) in past studies. Given that most previous Taq1A studies did not report C957T status, it is not possible to determine the effects of one SNP from another in those studies. We note, however, that despite the observed linkage disequilibrium, we only observed modest, nonsignificant effects for Taq1A in the present study, suggesting that linkage disequilibrium only partially accounts for past Taq1A findings.
Multilocus DRD2 score effects on D2/3 receptor availability
When probing for additional effects of Taq1A and Ins/Del to our observed main effects of C957T, we found little evidence for additional explanatory power for either SNP on our BPND effects. C957T alone accounted for most of the genetic variance in striatal BPND whether we focused our analyses on clusters identified from our multilocus score regression analyses or anatomically defined putamen and ventral striatum ROIs. However, we identified in our C957T+Ins/Del multilocus score analysis a midbrain/pons cluster (peak at 2, −26, −28, PFDR=0.087) not present in the C957T analysis alone. The location of this cluster ventral to the dopaminergic midbrain as well as the failure of the effect to reach significance when controlling for multiple comparisons make it difficult to draw conclusions about Ins/Del in this region (Supplementary Figure S1).76 We also observed a smaller cluster in midbrain/thalamus (k=144, at 8, −22, −4, PFDR=0.53). Given that variation in Fallypride BPND in midbrain and thalamus has been associated with schizophrenia,77 further investigation of genetic polymorphisms that affect BPND in these regions could aid in understanding risk for the disease. In addition, it is notable that individual differences in thalamic D2/3 receptor availability have been associated with differences in responses to dopaminergic drugs.78 Thus, genetic variants that affect DRD2 in the thalamus (or its subregions) may have implications for determining optimum pharmacological treatments. However, these extrastriatal findings should be interpreted with some caution until they are replicated.
C957T, BPND, and psychiatry
Our findings have implications for a variety of dopamine-linked psychiatric disorders. The C allele of C957T is more prevalent in patients with schizophrenia2, 3, 4, 18 and affects a variety of learning processes79, 80, 81, 82 as well as executive function.83, 84 However, despite the C957T effects observed here, differences in striatal D2/3 receptor availability (BPND) have not been consistently observed in contrasts of schizophrenics and healthy controls.77 This could reflect the difficulty of measuring D2/3 receptor levels in patients who may possess heightened DA synthesis capacity,85 which may impact both competition of radiotracers with endogenous dopamine,86, 87 and long-term regulation of D2/3 expression. Furthermore, additional short- and long-term impacts of antipsychotic medications on D2/3 receptor expression12, 14, 15 and dopamine regulation may impact PET measures in these patients. It is also conceivable that in the context of schizophrenia, C957T alters the impact of endogenous or exogenous perturbations of the dopamine system on D2/3 receptors. As such, it warrants particular attention in treatment research. Interestingly, the C allele has previously been associated with weight gain during treatment with antipsychotics.88
Furthermore, C957T has been associated with behavioral impulsivity,89, 90 whose effects increase with aging91 and reward sensitivity,92 which may explain why the C allele has been associated with increased risk for alcohol dependence.19 The lower D2/3 BPND we observed in the striatum of CC individuals fits with a wealth of data suggesting substance-dependent individuals display lower D2/3 BPND.93, 94 Furthermore our C957T BPND effects were strongest in the VS (accounting for 13 and 17% of the variance in right and left VS BPND, respectively), a key area involved in reward processing and dopamine release associated with drugs of abuse.95 There is some evidence that the C957T and Ins/Del SNPs predict quit rates in smokers treated with either bupropion or nicotine replacement therapy,96 illustrating the potential utility of using these genetic measures to target effective therapies. Although the mechanistic relationship between C957T and D2 receptor signaling still needs to be determined, the literature suggests that understanding this SNP (and others) may greatly aid us in treating individuals with a variety of psychiatric disorders.
Limitations
Although this study is larger than any other PET study to date examining gene effects on D2 receptor availability, we note that all such studies (including the present one) are underpowered. Although we used a common approach to index multilocus genetic effects45, 46, 47, 48 a more sophisticated approach, such as haplotype analysis, might offer additional insights, but require much larger samples due to the relative low minor allele frequency of the Taq1A (0.20) and -141C Ins/Del (0.09) SNPs. Furthermore, although we confirmed that our results remained consistent when controlling for ethnicity, it is possible that the results are stronger in particular ethnic groups, which we could not test for owing to small subject numbers with this further division of the data set. Indeed, the relative frequencies of the SNPs we investigated vary across ethnic groups68 and so the ethnic composition of this (69% Caucasian) and other studies warrants consideration when interpreting results. In addition, at least one study has suggested C957T effects may vary by individuals’ sex19 and although we controlled for it in our analyses, we were not well powered to test for sex by genotype interactions.
Conclusion
Our results replicate and extend previous work showing C957T T allele dosage is positively related to striatal D2/3 receptor availability (BPND) with significant effects observed in both the putamen and ventral striatum. Furthermore, we show that variation in this SNP explains a much larger portion of variability in striatal BPND than either the -141C Ins/Del or Taq1A alleles. By contrast, Taq1A alone or in combination with the other two tested DRD2 SNPs was not associated with striatal BPND, above the C957T effect. These findings demonstrate that DRD2 SNPs beyond Taq1A, specifically C957T, impact individual differences in striatal D2/3 BPND. To the extent that DRD2 relevant genes are interpreted as proxies in place of actual receptor assays, these data suggest that C957T is preferable to either -141C Ins/Del or Taq1A alleles.
References
Arinami T, Gao M, Hamaguchi H, Toru M . A functional polymorphism in the promoter region of the dopamine D2 receptor gene is associated with schizophrenia. Hum Mol Genet 1997; 6: 577–582.
Betcheva ET, Mushiroda T, Takahashi A, Kubo M, Karachanak SK, Zaharieva IT et al. Case-control association study of 59 candidate genes reveals the DRD2 SNP rs6277 (C957T) as the only susceptibility factor for schizophrenia in the Bulgarian population. J Hum Genet 2009; 54: 98–107.
Hanninen K, Katila H, Kampman O, Anttila S, Illi A, Rontu R et al. Association between the C957T polymorphism of the dopamine D2 receptor gene and schizophrenia. Neurosci Lett 2006; 407: 195–198.
Lawford BR, Young RM, Swagell CD, Barnes M, Burton SC, Ward WK et al. The C/C genotype of the C957T polymorphism of the dopamine D2 receptor is associated with schizophrenia. Schizophr Res 2005; 73: 31–37.
Zhang JP, Lencz T, Malhotra AK . D2 receptor genetic variation and clinical response to antipsychotic drug treatment: a meta-analysis. Am J Psychiatry 2010; 167: 763–772.
Lencz T, Robinson DG, Xu K, Ekholm J, Sevy S, Gunduz-Bruce H et al. DRD2 promoter region variation as a predictor of sustained response to antipsychotic medication in first-episode schizophrenia patients. Am J Psychiatry 2006; 163: 529–531.
Zhang JP, Robinson DG, Gallego JA, John M, Yu J, Addington J et al. Association of a schizophrenia risk variant at the DRD2 locus with antipsychotic treatment response in first-episode psychosis. Schizophr Bull 2015; 41: 1248–1255.
Reynolds GP, McGowan OO, Dalton CF . Pharmacogenomics in psychiatry: the relevance of receptor and transporter polymorphisms. Br J Clin Pharmacol 2014; 77: 654–672.
Arranz MJ, de Leon J . Pharmacogenetics and pharmacogenomics of schizophrenia: a review of last decade of research. Mol Psychiatry 2007; 12: 707–747.
Creese I, Burt DR, Snyder SH . Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science 1976; 192: 481–483.
Seeman P, Lee T, Chau-Wong M, Wong K . Antipsychotic drug doses and neuroleptic/dopamine receptors. Nature 1976; 261: 717–719.
Farde L, Wiesel FA, Nordstrom AL, Sedvall G . D1- and D2-dopamine receptor occupancy during treatment with conventional and atypical neuroleptics. Psychopharmacology 1989; S28–S31.
Howes OD, Egerton A, Allan V, McGuire P, Stokes P, Kapur S . Mechanisms underlying psychosis and antipsychotic treatment response in schizophrenia: insights from PET and SPECT imaging. Curr Pharm Des 2009; 15: 2550–2559.
Kapur S, Zipursky R, Jones C, Remington G, Houle S . Relationship between dopamine D(2) occupancy, clinical response, and side effects: a double-blind PET study of first-episode schizophrenia. Am J Psychiatry 2000; 157: 514–520.
Nordstrom AL, Farde L, Wiesel FA, Forslund K, Pauli S, Halldin C et al. Central D2-dopamine receptor occupancy in relation to antipsychotic drug effects: a double-blind PET study of schizophrenic patients. Biol Psychiatry 1993; 33: 227–235.
Stockmeier CA, DiCarlo JJ, Zhang Y, Thompson P, Meltzer HY . Characterization of typical and atypical antipsychotic drugs based on in vivo occupancy of serotonin2 and dopamine2 receptors. J Pharm Exp Ther 1993; 266: 1374–1384.
Stone JM, Davis JM, Leucht S, Pilowsky LS . Cortical dopamine D2/D3 receptors are a common site of action for antipsychotic drugs—an original patient data meta-analysis of the SPECT and PET in vivo receptor imaging literature. Schizophr Bull 2009; 35: 789–797.
Liu L, Fan D, Ding N, Hu Y, Cai G, Wang L et al. The relationship between DRD2 gene polymorphisms (C957T and C939T) and schizophrenia: a meta-analysis. Neurosci Lett 2014; 583: 43–48.
Swagell CD, Lawford BR, Hughes IP, Voisey J, Feeney GF, van Daal A et al. DRD2 C957T and TaqIA genotyping reveals gender effects and unique low-risk and high-risk genotypes in alcohol dependence. Alcohol Alcohol 2012; 47: 397–403.
Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet 2006; 15: 3498–3507.
Hill SY, Hoffman EK, Zezza N, Thalamuthu A, Weeks DE, Matthews AG et al. Dopaminergic mutations: within-family association and linkage in multiplex alcohol dependence families. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 517–526.
Hirvonen M, Laakso A, Nagren K, Rinne J, Pohjalainen T, Hietala J . Corrigendum: C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry 2005; 10: 889.
Hirvonen M, Laakso A, Nagren K, Rinne JO, Pohjalainen T, Hietala J . C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry 2004; 9: 1060–1061.
Hirvonen MM, Lumme V, Hirvonen J, Pesonen U, Nagren K, Vahlberg T et al. C957T polymorphism of the human dopamine D2 receptor gene predicts extrastriatal dopamine receptor availability in vivo. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33: 630–636.
Jonsson EG, Nothen MM, Grunhage F, Farde L, Nakashima Y, Propping P et al. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatry 1999; 4: 290–296.
Pohjalainen T, Nagren K, Syvalahti EK, Hietala J . The dopamine D2 receptor 5'-flanking variant, -141C Ins/Del, is not associated with reduced dopamine D2 receptor density in vivo. Pharmacogenetics 1999; 9: 505–509.
Pohjalainen T, Rinne JO, Nagren K, Lehikoinen P, Anttila K, Syvalahti EK et al. The A1 allele of the human D2 dopamine receptor gene predicts low D2 receptor availability in healthy volunteers. Mol Psychiatry 1998; 3: 256–260.
Thompson J, Thomas N, Singleton A, Piggott M, Lloyd S, Perry EK et al. D2 dopamine receptor gene (DRD2) Taq1 A polymorphism: reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics 1997; 7: 479–484.
Laruelle M, Gelernter J, Innis RB . D2 receptors binding potential is not affected by Taq1 polymorphism at the D2 receptor gene. Mol Psychiatry 1998; 3: 261–265.
Brody AL, Mandelkern MA, Olmstead RE, Scheibal D, Hahn E, Shiraga S et al. Gene variants of brain dopamine pathways and smoking-induced dopamine release in the ventral caudate/nucleus accumbens. Arch Gen Psychiatry 2006; 63: 808–816.
Wagner AK, Scanlon JM, Becker CR, Ritter AC, Niyonkuru C, Dixon CE et al. The influence of genetic variants on striatal dopamine transporter and D2 receptor binding after TBI. J Cereb Blood Flow Metab 2014; 34: 1328–1339.
Jocham G, Klein TA, Neumann J, von Cramon DY, Reuter M, Ullsperger M . Dopamine DRD2 polymorphism alters reversal learning and associated neural activity. J Neurosci 2009; 29: 3695–3704.
Klein TA, Neumann J, Reuter M, Hennig J, von Cramon DY, Ullsperger M . Genetically determined differences in learning from errors. Science 2007; 318: 1642–1645.
Richter A, Guitart-Masip M, Barman A, Libeau C, Behnisch G, Czerney S et al. Valenced action/inhibition learning in humans is modulated by a genetic variant linked to dopamine D2 receptor expression. Front Syst Neurosci 2014; 8: 140.
Richter A, Richter S, Barman A, Soch J, Klein M, Assmann A et al. Motivational salience and genetic variability of dopamine D2 receptor expression interact in the modulation of interference processing. Front Hum Neurosci 2013; 7: 250.
Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA 2007; 104: 20552–20557.
Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 2003; 12: 205–216.
Stelzel C, Basten U, Montag C, Reuter M, Fiebach CJ . Frontostriatal involvement in task switching depends on genetic differences in d2 receptor density. J Neurosci 2010; 30: 14205–14212.
Forbes EE, Brown SM, Kimak M, Ferrell RE, Manuck SB, Hariri AR . Genetic variation in components of dopamine neurotransmission impacts ventral striatal reactivity associated with impulsivity. Mol Psychiatry 2009; 14: 60–70.
Cohen MX, Young J, Baek JM, Kessler C, Ranganath C . Individual differences in extraversion and dopamine genetics predict neural reward responses. Brain Res Cogn Brain Res 2005; 25: 851–861.
Frank MJ, Fossella JA . Neurogenetics and pharmacology of learning, motivation, and cognition. Neuropsychopharmacology 2011; 36: 133–152.
Gluskin BS, Mickey BJ . Genetic variation and dopamine D2 receptor availability: a systematic review and meta-analysis of human in vivo molecular imaging studies. Transl Psychiatry 2016; 6: e747.
Davis C, Loxton NJ . Addictive behaviors and addiction-prone personality traits: associations with a dopamine multilocus genetic profile. Addict Behav 2013; 38: 2306–2312.
Davis C, Loxton NJ, Levitan RD, Kaplan AS, Carter JC, Kennedy JL . 'Food addiction' and its association with a dopaminergic multilocus genetic profile. Physiol Behav 2013; 118: 63–69.
Nikolova YS, Ferrell RE, Manuck SB, Hariri AR . Multilocus genetic profile for dopamine signaling predicts ventral striatum reactivity. Neuropsychopharmacology 2011; 36: 1940–1947.
Stice E, Yokum S, Burger K, Epstein L, Smolen A . Multilocus genetic composite reflecting dopamine signaling capacity predicts reward circuitry responsivity. J Neurosci 2012; 32: 10093–10100.
Yokum S, Marti CN, Smolen A, Stice E . Relation of the multilocus genetic composite reflecting high dopamine signaling capacity to future increases in BMI. Appetite 2015; 87: 38–45.
Kohno M, Nurmi EL, Laughlin CP, Morales AM, Gail EH, Hellemann GS et al. Functional genetic variation in dopamine signaling moderates prefrontal cortical activity during risky decision making. Neuropsychopharmacology 2016; 41: 695–703.
Zald DH, Woodward ND, Cowan RL, Riccardi P, Ansari MS, Baldwin RM et al. The interrelationship of dopamine D2-like receptor availability in striatal and extrastriatal brain regions in healthy humans: a principal component analysis of [18F]fallypride binding. Neuroimage 2010; 51: 53–62.
First MB . Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Patient Edition. Biometrics Research Department Columbia University: New York, NY, USA, 2005.
Smith CT, Dang LC, Cowan RL, Kessler RM, Zald DH . Variability in paralimbic dopamine signaling correlates with subjective responses to d-amphetamine. Neuropharmacology 2016; 108: 394–402.
Friston KJ, Holmes AP, Worsley KJ, Poline J-P, Frith CD, Frackowiak RSJ . Statistical parametic maps in functional imaging: a general linear approach. Hum Brain Mapp 1995; 2: 189–210.
Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004; 23 (Suppl 1): S208–S219.
Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH . An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage 2003; 19: 1233–1239.
Lammertsma AA, Hume SP . Simplified reference tissue model for PET receptor studies. Neuroimage 1996; 4 (3 Pt 1): 153–158.
Gunn RN, Lammertsma AA, Hume SP, Cunningham VJ . Parametric imaging of ligand-receptor binding in PET using a simplified reference region model. Neuroimage 1997; 6: 279–287.
Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010; 86: 560–572.
Matuskey D, Worhunksy P, Correa E, Pittman B, Gallezot JD, Nabulsi N et al. Age-related changes in binding of the D2/3 receptor radioligand [(11)C](+)PHNO in healthy volunteers. Neuroimage 2016; 130: 241–247.
Mukherjee J, Christian BT, Dunigan KA, Shi B, Narayanan TK, Satter M et al. Brain imaging of 18F-fallypride in normal volunteers: blood analysis, distribution, test-retest studies, and preliminary assessment of sensitivity to aging effects on dopamine D-2/D-3 receptors. Synapse 2002; 46: 170–188.
Pohjalainen T, Rinne JO, Nagren K, Syvalahti E, Hietala J . Sex differences in the striatal dopamine D2 receptor binding characteristics in vivo. Am J Psychiatry 1998; 155: 768–773.
Riccardi P, Zald D, Li R, Park S, Ansari MS, Dawant B et al. Sex differences in amphetamine-induced displacement of [(18)F]fallypride in striatal and extrastriatal regions: a PET study. Am J Psychiatry 2006; 163: 1639–1641.
Savitz J, Hodgkinson CA, Martin-Soelch C, Shen PH, Szczepanik J, Nugent AC et al. DRD2/ANKK1 Taq1A polymorphism (rs1800497) has opposing effects on D2/3 receptor binding in healthy controls and patients with major depressive disorder. Int J Neuropsychopharmacol 2013; 16: 2095–2101.
Mawlawi O, Martinez D, Slifstein M, Broft A, Chatterjee R, Hwang DR et al. Imaging human mesolimbic dopamine transmission with positron emission tomography: I. Accuracy and precision of D(2) receptor parameter measurements in ventral striatum. J Cereb Blood Flow Metab 2001; 21: 1034–1057.
Dang LC, O'Neil JP, Jagust WJ . Genetic effects on behavior are mediated by neurotransmitters and large-scale neural networks. Neuroimage 2012; 66C: 203–214.
Dang LC, O'Neil JP, Jagust WJ . Dopamine supports coupling of attention-related networks. J Neurosci 2012; 32: 9582–9587.
Dang LC, Samanez-Larkin GR, Castrellon JJ, Perkins SF, Cowan RL, Zald DH . Associations between dopamine D2 receptor availability and BMI depend on age. Neuroimage 2016; 138: 176–183.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–289.
International HapMap Consortium. A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Monsma FJ Jr., Barton AC, Sibley DR . Expression of functional D2 dopamine receptors following differentiation of Y-79 human retinoblastoma cells. J Neurochem 1990; 54: 1200–1207.
Morris ED, Yoder KK . Positron emission tomography displacement sensitivity: predicting binding potential change for positron emission tomography tracers based on their kinetic characteristics. J Cereb Blood Flow Metab 2007; 27: 606–617.
Slifstein M, Kegeles LS, Xu X, Thompson JL, Urban N, Castrillon J et al. Striatal and extrastriatal dopamine release measured with PET and [(18)F] fallypride. Synapse 2010; 64: 350–362.
Laakso A, Pohjalainen T, Bergman J, Kajander J, Haaparanta M, Solin O et al. The A1 allele of the human D2 dopamine receptor gene is associated with increased activity of striatal L-amino acid decarboxylase in healthy subjects. Pharmacogenet Genomics 2005; 15: 387–391.
Eisenstein SA, Bogdan R, Love-Gregory L, Corral-Frias NS, Koller JM, Black KJ et al. Prediction of striatal D2 receptor binding by DRD2/ANKK1 TaqIA allele status. Synapse 2016; 70: 418–431.
Machiela MJ, Chanock SJ . LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015; 31: 3555–3557.
Willeit M, Praschak-Rieder N . Imaging the effects of genetic polymorphisms on radioligand binding in the living human brain: a review on genetic neuroreceptor imaging of monoaminergic systems in psychiatry. Neuroimage 2010; 53: 878–892.
Murty VP, Shermohammed M, Smith DV, Carter RM, Huettel SA, Adcock RA . Resting state networks distinguish human ventral tegmental area from substantia nigra. Neuroimage 2014; 100: 580–589.
Kessler RM, Woodward ND, Riccardi P, Li R, Ansari MS, Anderson S et al. Dopamine D2 receptor levels in striatum, thalamus, substantia nigra, limbic regions, and cortex in schizophrenic subjects. Biol Psychiatry 2009; 65: 1024–1031.
Samanez-Larkin GR, Buckholtz JW, Cowan RL, Woodward ND, Li R, Ansari MS et al. A thalamocorticostriatal dopamine network for psychostimulant-enhanced human cognitive flexibility. Biol Psychiatry 2013; 74: 99–105.
Frank MJ, Hutchison K . Genetic contributions to avoidance-based decisions: striatal D2 receptor polymorphisms. Neuroscience 2009; 164: 131–140.
Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE . Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proc Natl Acad Sci USA 2007; 104: 16311–16316.
Huertas E, Buhler KM, Echeverry-Alzate V, Gimenez T, Lopez-Moreno JA . C957T polymorphism of the dopamine D2 receptor gene is associated with motor learning and heart rate. Genes Brain Behav 2012; 11: 677–683.
Xie Z, Maddox WT, McGeary JE, Chandrasekaran B . The C957T polymorphism in the dopamine receptor D(2) gene modulates domain-general category learning. J Neurophysiol 2015; 113: 3281–3290.
Rodriguez-Jimenez R, Hoenicka J, Jimenez-Arriero MA, Ponce G, Bagney A, Aragues M et al. Performance in the Wisconsin Card Sorting Test and the C957T polymorphism of the DRD2 gene in healthy volunteers. Neuropsychobiology 2006; 54: 166–170.
Xu H, Kellendonk CB, Simpson EH, Keilp JG, Bruder GE, Polan HJ et al. DRD2 C957T polymorphism interacts with the COMT Val158Met polymorphism in human working memory ability. Schizophr Res 2007; 90: 104–107.
Howes OD, Kambeitz J, Kim E, Stahl D, Slifstein M, Abi-Dargham A et al. The nature of dopamine dysfunction in schizophrenia and what this means for treatment. Arch Gen Psychiatry 2012; 69: 776–786.
Laruelle M, D'Souza CD, Baldwin RM, Abi-Dargham A, Kanes SJ, Fingado CL et al. Imaging D2 receptor occupancy by endogenous dopamine in humans. Neuropsychopharmacology 1997; 17: 162–174.
Laruelle M, Huang Y . Vulnerability of positron emission tomography radiotracers to endogenous competition. New insights. Q J Nucl Med 2001; 45: 124–138.
Muller DJ, Zai CC, Sicard M, Remington E, Souza RP, Tiwari AK et al. Systematic analysis of dopamine receptor genes (DRD1-DRD5) in antipsychotic-induced weight gain. Pharmacogenomics J 2012; 12: 156–164.
Colzato LS, van den Wildenberg WP, Van der Does AJ, Hommel B . Genetic markers of striatal dopamine predict individual differences in dysfunctional, but not functional impulsivity. Neuroscience 2010; 170: 782–788.
White MJ, Lawford BR, Morris CP, Young RM . Interaction between DRD2 C957T polymorphism and an acute psychosocial stressor on reward-related behavioral impulsivity. Behav Genet 2009; 39: 285–295.
Colzato LS, van den Wildenberg WP, Hommel B . The genetic impact (C957T-DRD2) on inhibitory control is magnified by aging. Neuropsychologia 2013; 51: 1377–1381.
Davis C, Levitan RD, Kaplan AS, Carter J, Reid C, Curtis C et al. Reward sensitivity and the D2 dopamine receptor gene: A case-control study of binge eating disorder. Prog Neuropsychopharmacol Biol Psychiatry 2008; 32: 620–628.
Martinez D, Broft A, Foltin RW, Slifstein M, Hwang DR, Huang Y et al. Cocaine dependence and d2 receptor availability in the functional subdivisions of the striatum: relationship with cocaine-seeking behavior. Neuropsychopharmacology 2004; 29: 1190–1202.
Volkow ND, Fowler JS, Wang GJ, Swanson JM . Dopamine in drug abuse and addiction: results from imaging studies and treatment implications. Mol Psychiatry 2004; 9: 557–569.
Pierce RC, Kumaresan V . The mesolimbic dopamine system: the final common pathway for the reinforcing effect of drugs of abuse? Neurosci Biobehav Rev 2006; 30: 215–238.
Lerman C, Jepson C, Wileyto EP, Epstein LH, Rukstalis M, Patterson F et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology 2006; 31: 231–242.
Acknowledgements
This work was supported by the National Institute on Drug Abuse (R01DA019670 and R21DA033611 to DHZ; F32DA041157 to CTS; F32DA036979 to LCD), National Institute on Aging (R01AG044848 to DHZ), and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Smith, C., Dang, L., Buckholtz, J. et al. The impact of common dopamine D2 receptor gene polymorphisms on D2/3 receptor availability: C957T as a key determinant in putamen and ventral striatum. Transl Psychiatry 7, e1091 (2017). https://doi.org/10.1038/tp.2017.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2017.45
This article is cited by
-
Blocking D2/D3 dopamine receptors in male participants increases volatility of beliefs when learning to trust others
Nature Communications (2023)
-
Effects of OPRM1 and DRD2 on brain structure in drug-naïve adolescents: Genetic and neural vulnerabilities to substance use
Psychopharmacology (2022)
-
Cortical thickness mediates the relationship between DRD2 C957T polymorphism and executive function across the adult lifespan
Brain Structure and Function (2021)
-
Lack of consistent sex differences in d-amphetamine-induced dopamine release measured with [18F]fallypride PET
Psychopharmacology (2019)
-
Association of genetic ancestry with striatal dopamine D2/D3 receptor availability
Molecular Psychiatry (2018)