Abstract
Mild cognitive impairment (MCI) is considered as a transition phase between normal aging and Alzheimer's disease (AD). MCI confers an increased risk of developing AD, although the state is heterogeneous with several possible outcomes, including even improvement back to normal cognition. We sought to determine the serum metabolomic profiles associated with progression to and diagnosis of AD in a prospective study. At the baseline assessment, the subjects enrolled in the study were classified into three diagnostic groups: healthy controls (n=46), MCI (n=143) and AD (n=47). Among the MCI subjects, 52 progressed to AD in the follow-up. Comprehensive metabolomics approach was applied to analyze baseline serum samples and to associate the metabolite profiles with the diagnosis at baseline and in the follow-up. At baseline, AD patients were characterized by diminished ether phospholipids, phosphatidylcholines, sphingomyelins and sterols. A molecular signature comprising three metabolites was identified, which was predictive of progression to AD in the follow-up. The major contributor to the predictive model was 2,4-dihydroxybutanoic acid, which was upregulated in AD progressors (P=0.0048), indicating potential involvement of hypoxia in the early AD pathogenesis. This was supported by the pathway analysis of metabolomics data, which identified upregulation of pentose phosphate pathway in patients who later progressed to AD. Together, our findings primarily implicate hypoxia, oxidative stress, as well as membrane lipid remodeling in progression to AD. Establishment of pathogenic relevance of predictive biomarkers such as ours may not only facilitate early diagnosis, but may also help identify new therapeutic avenues.
Similar content being viewed by others
Introduction
Alzheimer's disease (AD) is a growing challenge to the health care systems and economies of developed countries, with millions of patients suffering from this disease and increasing numbers of new cases diagnosed annually with the increasing age of populations.1 Mild cognitive impairment (MCI) is considered as a transition phase between normal aging and AD.2 A subject with MCI shows cognitive impairment, primarily in memory functions, yet has preserved activities of daily living and does not fulfill the criteria of AD or any other dementia disorder. MCI confers an increased risk of developing AD,3 although the state is heterogeneous with several possible outcomes, including even improvement back to normal cognition.4 Recent research has thus concentrated on obtaining biomarkers to identify features that differentiate between those MCI subjects who will develop AD (progressive MCI, P-MCI) from stable MCI (S-MCI) and healthy elderly control subjects.
Ideally, the AD biomarkers (1) would reflect the disease-related biological processes and (2) may be measured non-invasively, such as a blood test. The molecular markers sensitive to the underlying pathogenic factors would be of high relevance not only to assist early disease detection and diagnosis, but also to subsequently facilitate the disease monitoring and treatment responses. Promising, although non-overlapping, results have been obtained in two independent plasma proteomics studies aiming to identify potential markers predictive of AD.5, 6 Metabolomics is a discipline dedicated to the global study of small molecules (i.e., metabolites) in cells, tissues and biofluids. Concentration changes of specific groups of metabolites may be sensitive to pathogenically relevant factors such as genetic variation,7 diet,8 age,9, 10 immune system status11 or gut microbiota,12 and their study may therefore be a powerful tool for characterization of complex phenotypes affected by both genetic and environmental factors.13 In the past years, technologies have been developed that allow comprehensive and quantitative investigation of a multitude of different metabolites.14
Among the metabolites, lipids have received most attention, as all amyloid precursor protein-processing proteins are transmembrane proteins.15 Lipids are major constituents of cell membranes, and their composition is important to maintain membrane fluidity, topology, mobility or activity of membrane-bound proteins, and to ensure normal cellular physiology.16 Investigations of disease-related ‘lipidome’ covering a global profile of structurally and functionally diverse lipids provide an opportunity to pursue, accurately and sensitively, studies profiling hundreds of molecular lipids in parallel.17, 18 The so-called lipidomics approach may not only provide information about the disease-related markers, but in addition, deliver clues about the mechanisms behind the control of cellular lipid homeostasis.16
Herein, we sought to determine the serum metabolomic profiles associated with progression to and diagnosis of AD in a well-characterized prospective study. At the baseline assessment, subjects enrolled in the study were classified into three diagnostic groups: healthy controls, MCI and AD. Global metabolomics approach using two platforms with broad analytical coverage, from lipids to small polar metabolites, was applied to analyze baseline serum samples from subjects involved in the study, and to associate the metabolite profiles with the diagnosis at the baseline and in the follow-up.
Methods
Participants
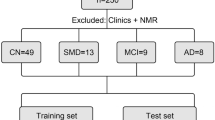
Within the PredictAD project (http://www.predictad.eu/), focusing on predictors of conversion of MCI to clinical AD dementia, 143 subjects diagnosed with MCI were pooled from longitudinal study databases gathered in the University of Kuopio, and their findings were compared with those of 46 healthy control subjects and 37 AD patients.19, 20, 21 The blood samples were taken during morning hours and after fasting in most cases. A venous blood sample was collected into heparin tubes and plasma was separated using standard methods. The samples were aliquoted and stored in polypropylene tubes at −70 °C until analyses. Descriptive and clinical data of the study groups are presented in Table 1.
Healthy control subjects included in this study were volunteers from the population-based cohorts, and the methods used for the identification of control subjects have been described in previous studies.19, 20 They had no history of neurological or psychiatric diseases and showed no impairment in the detailed neuropsychological evaluation.
MCI was diagnosed using the criteria originally proposed by the Mayo Clinic Alzheimer's Disease Research Center.22, 23 These criteria have later been modified, but at the time this study population was recruited, the MCI criteria required were as follows: (1) memory complaint by patient, family or physician; (2) normal activities of daily living; (3) normal global cognitive function; (4) objective impairment in memory or in one other area of cognitive function as evident by scores >1.5 s.d. below the age-appropriate mean; (5) Clinical Dementia Rating (CDR) score of 0.5; and (6) absence of dementia. As the subjects were pooled from different study databases with slightly different neuropsychological test batteries, two scales, which were done with all the MCI subjects, were selected to describe their cognitive status, mini-mental state examination (MMSE) and CDR sum of boxes. Although the neuropsychological test battery used to diagnose MCI varied slightly, all the MCI subjects were considered having the amnestic subtype of the syndrome at the time of recruitment.
Diagnosis of AD included evaluation of medical history, physical and neurological examinations performed by a physician, and a detailed neuropsychological evaluation. The severity of the cognitive decline was graded according to the CDR Scale.24 Brain magnetic resonance imaging scan, cerebrospinal fluid (CSF) analysis, electrocardiography, chest radiography, screening for hypertension and depression, and blood tests were also performed to exclude other possible pathologies underlying the symptoms. The diagnosis of dementia was based on the criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th edition25 and the diagnosis of AD on the National Institute of Neurologic and Communicative Disorders and Stroke, and Alzheimer's Disease and Related Disorders Association criteria.26 All magnetic resonance images were also read by an experienced neuroradiologist to exclude subjects with severe white matter lesions or other abnormalities. The study subjects with a history of stroke or transient ischemic attack were excluded and accordingly, subjects with extensive confluent white matter lesions.
MCI subjects who developed AD during the course of the follow-up were considered as P-MCI subjects (n=52) and those whose status remained stable or improved (i.e., those who were later diagnosed as controls) were considered having S-MCI (n=91). The follow-up time for the P-MCI subjects (27±18 months, Table 1) was set to start at the baseline date and considered completed at the time of AD diagnosis. In the case of S-MCI subjects, the follow-up time (28±16 months, Table 1) was calculated as the time from baseline date to the last available evaluation date. For all subjects magnetic resonance images were acquired with 1.5 T magnetic resonance imaging scan in the Department of Clinical Radiology, Kuopio University Hospital.27 The Apolipoprotein E (APOE) genotype of the study subjects was determined by using a standard protocol.28 The APOE allelic distribution within the study groups is presented in Table 1.
Informed written consent was acquired from all the subjects according to the Declaration of Helsinki, and the study was approved by the Ethics Committee of Kuopio University Hospital.
Metabolomic analysis
Two analytical platforms for metabolomics were applied to all samples from the estimation cohort: (1) global lipidomics platform, based on ultra performance liquid chromatography coupled to mass spectrometry (MS), covers molecular lipids such as phospholipids, sphingolipids and neutral lipids; (2) platform for global profiling of small polar metabolites, based on comprehensive two-dimensional gas chromatography coupled to time-of-flight mass spectrometry (GC × GC–TOFMS), covers small molecules such as amino acids, free fatty acids, keto-acids, various other organic acids, sterols and sugars. Both platforms were recently described in detail29, 30 and are also described in Supplementary Methods. Raw ultra performance liquid chromatography coupled to MS and GC × GC–TOFMS data were processed with MZmine 231 and guineu30 software, respectively. The final data set from each platform consisted of a list of metabolite peaks (identified or unidentified) and their levels, calculated using the platform-specific methods, across all samples. All metabolite peaks were included in the data analyses, including the unidentified ones. We reasoned that inclusion of complete data as obtained from the platform best represents the global metabolome, and the unidentified peaks may still be followed-up later on with de novo identification, using additional experiments if considered of interest.
Descriptive statistical analyses
Statistical analyses for clinical data were performed by SPSS software release 14.0.1 for Windows (SPSS, Chicago, IL, USA). The comparisons between the different study groups were done by independent samples t-test. Otherwise, if the assumptions for normality were not met, the non-parametric tests were used. For the categorical data, the comparisons between different groups were made using the χ2-tests.
One-way analysis of variance (ANOVA), implemented in Matlab (MathWorks, Natick, MA, USA), was applied to compare the average within-cluster metabolite profiles between the diagnostic groups. The statistical analyses at individual metabolite level were performed using R version 2.13. The median values of metabolites across the three diagnostic groups at baseline were compared using the Kruskal–Wallis one-way ANOVA, whereas the medians of P-MCI and S-MCI groups were compared by Wilcoxon test. Individual metabolite levels were visualized using the beanplots,32 implemented in ‘beanplot’ R package. Beanplot provides information on the mean metabolite level within each group, density of the data-point distribution, as well as shows individual data points.
Cluster analysis
The data were scaled to zero mean and unit variance, to obtain metabolite profiles comparable to each other. Bayesian model-based clustering was applied on the scaled data to group lipids, which were similarly expressed across all samples. The analyses were performed using MCLUST33 method, implemented in R statistical language34 as package ‘mclust’. In MCLUST, the observed data are viewed as a mixture of several clusters and each cluster comes from a unique probability density function. A number of clusters in the mixture, together with the cluster-specific parameters that constrain the probability distributions, will define a model, which can then be compared with others. The clustering process selects the optimal model and determines the data partition accordingly.
A number of clusters ranging from 4–15 and all available model families were considered in our study. Models were compared using the Bayesian information criterion, which is an approximation of the marginal likelihood. The best model is the one that gives the largest marginal likelihood of data, that is, the highest Bayesian information criterion value.
Diagnostic model
The best marker combination was searched for in two phases: in the first phase, penalized, generalized linear models35 were used to pre-screen a prominent marker set, and in the second phase, a step-wise optimization algorithm was used to optimize the marker combination. In both phases, 1000 cross-validation runs were performed. In each run, two out of three and one out of three of the samples were selected at random to the training and test sets, respectively. In the first phase, markers leading to lowest coefficient of variation errors were selected.
In the second phase, logistic regression model implemented in R was applied to discriminate the groups of interest. The best marker combination in the logistic regression model was selected by step-wise algorithm using Akaike's information criterion.36 The best model was then applied to the test set samples to calculate their predicted classes. The optimal marker combinations in each of the cross-validation runs, receiver-operating characteristic curves with area under the curve (AUC) statistics, odds ratios and relative risks were recorded. Different biomarker signatures were then compared on the basis of the number of times they were selected as the best performing models. The performance of the top-ranking signature was then reported using the same procedure as above, but only considering the selected combination of metabolites. Receiver-operating characteristic curves with AUC statistics, prediction accuracy, odds ratios and relative risks were recorded on the basis of the performance in the independently tested data (one out of three of the samples) for each of the 2000 cross-validation runs.
Different models, for example, model based on metabolites alone versus model based on APOE genotype, as well as metabolites, were compared using the likelihood ratio test, which expresses how many times more likely the data are under one model than the other to compare their fit with the data.37
Pathway analysis
MPEA (metabolic pathway enrichment analysis)38 is a tool for functional analysis and biological interpretation of metabolic profiling data generated by GC–MS. The concept of MPEA is the same as that of widely-accepted gene set enrichment analysis.39 MPEA accepts a ranked list of mass spectra and tests whether metabolites belonging to some metabolic pathway tend to occur toward the top (or bottom) of this ranked mass chromatogram. Herein, MPEA was applied using the default parameters: permutations=100, kselection=1, penalty_mode=0, organism=HSA, gsea=1, direction=2, list_size=all, column=VAR5, column_width=25, dotproduct=0.05, euclideandist=0.05, hammingdist=50, jaccarddist=0.6, binarydist=0.6.
Results
Metabolomics in a prospective cohort
Using the two analytical platforms, a total of 139 molecular lipids and 544 small polar metabolites were measured, respectively, from 226 serum samples (Table 1). Due to a high degree of co-regulation among the metabolites,40 one cannot assume that all the 683 measured metabolites are independent. For this reason the global metabolome was first surveyed by clustering the data into a subset of clusters, using the Bayesian model-based clustering.33 Such an approach decomposes the metabolome into specific clusters of co-varying metabolites. The so-obtained clusters and their average levels across different sample groups provide a global view of the main metabolic changes. As a potential disadvantage, such analysis may miss potentially interesting outlier metabolites, which greatly vary between the diagnostic groups, but are not well represented by any of the average cluster profiles.
Lipidomic platform data was decomposed into seven (LCs) and the GC × GC—TOFMS-based metabolomic data into six clusters (MCs), respectively
Description of each cluster and the representative metabolites are shown in Table 2. As expected, the division of clusters to a large extent follows different metabolite functional or structural groups. As shown in Figure 1a (and in Figure 1b for selected representative identified metabolites), several of the clusters had different average metabolite profiles across the three diagnostic groups at the baseline. Specifically, there was an overall trend towards lower lipid levels in AD, with the highest levels in the control group (LCs 3–7). The differences of average within-cluster profiles between the three groups reached the significance level in LC1, LC3 (both containing predominantly phosphatidylcholines (PC)) and LC4 (consisting predominantly of ether phospholipids, including plasmalogens). When corrected for age and APOE genotype, only the LC4 remained statistically significant, whereas LC1 was marginally significant (P=0.07). Among the metabolites, MC3 was different between the diagnostic groups at baseline at a marginal significance level, but was not significant after correction for age and APOE genotype. The two large clusters, MC1 and MC2, did not change on average between the groups, but did contain several significantly changing metabolites.
Metabolomic profiles across the three diagnostic groups at baseline. (a) Mean metabolite levels within each cluster. Error marks show s.e.m. (*P<0.05). When correcting for age and ApoE genotype, only LC4 remained statistically significant, whereas LC1 was marginally significant (P=0.07). (b) Profiles of selected representative metabolites from different clusters in control and Alzheimer's disease (AD) groups at baseline. The metabolite levels are shown as beanplots,32 which provide information on the mean level (solid line), individual data points (short lines), and the density of the distribution. The concentration scale in beanplots is logarithmic for some metabolites.
Feasibility of diagnosis and prediction of AD
To assess the feasibility of diagnosis, we performed a model selection in multiple-cross validation runs as described in the ‘Methods’ section. The best model derived from logistic regression analysis was obtained by combining four metabolites: two PC (PC (18:0/18:2) from LC1 and PC (16:0/20:4) from LC5), lactic acid (MC2; PubChem CID 61503) and ketovaline (MC3; PubChem CID 49). This combination was selected in 248 out of 1000 cross-validation runs. The next three strongly performing models, which were together selected in 275 out of 1000 cross-validation runs, were closely related, as they contained the subsets of two or three metabolites of the top-ranking model. The model performed reasonably well, with AUC=0.77, 90% CI=(0.66, 0.88). Sensitivity and specificity on the basis of optimal cut-off point were 0.64, 90% CI=(0.40, 0.85) and 0.72, 90% CI=(0.56, 0.86), respectively. Supplementary Figure S1 shows the receiver-operating characteristic curve of the diagnostic model comprising the four metabolites, based on the independently tested data taken from 2000 samplings.
We also included age and APOE genotype (APOE ɛ4 genotype present or absent) in the diagnostic model. APOE or age alone performed worse than metabolic signature (P<0.001). For the model based on APOE genotype alone, AUC=0.61, 90% CI=(0.49, 0.73; Supplementary Figure 2). Combining metabolic signature and APOE genotype did not improve the model (P=0.48) (Supplementary Figure 3). However, combining age alone, or age and APOE genotype together with the metabolic signature did improve the model (P=0.006 and P=0.019, respectively; Supplementary Figures 4 and 5). The best performing model was based on the metabolite signature together with age, with AUC=0.81, 90% CI=(0.69, 0.91), sensitivity of 0.67, 90% CI=(0.44, 0.90) and specificity of 0.76, 90% CI=(0.60, 0.89).
We also tested if any of the patients in the progressive MCI group had the AD metabolic profile. When applying the AD versus control group classification to the P-MCI group, 12 MCI patients (24%) who later progressed to AD were identified as having the AD metabolic signature.
We then investigated the feasibility of prediction of AD by comparing stable and progressive MCI groups on the basis of metabolomics profiles at baseline. Using the same approach as above, the best model contained three metabolites: PC from LC3 (PC (16:0/16:0)), an unidentified carboxylic acid (MC2) and 2,4-dihydroxybutanoic acid (MC1; PubChem CID 192742). The top model was selected in 195 out of 1000 cross-validation runs. Other best-selected models contained the two metabolites (carboxylic acid and 2,4-dihydroxybutanoic acid), but with varying lipids (including lysoPC (16:0), PC (16:0/20:5), PC (18:0/20:4) or PC (O-18:1/16:0)), or without.
The metabolic signature obtained predicted AD reasonably well, with AUC=0.77, 90% CI=(0.65, 0.87), sensitivity of 0.77, 90% CI=(0.53, 1.00), specificity of 0.70, 90% CI=(0.53, 0.86) and odds ratio of 8.0, 90% CI=(2.7, 27.6). Figure 2 shows the receiver-operating characteristic curve of the combined diagnostic model comprising three metabolites, based on the independently tested data taken from 2000 samplings. Supplementary Figure 6 shows the levels of the three metabolites included in the biomarker across all four patient groups included in the study. Interestingly, the increase of 2,4-dihydroxybutanoic acid concentration appears to be specific to the P-MCI group, whereas none of the metabolites display the progressive changes from healthy controls to AD.
Feasibility of predicting Alzheimer's disease (AD), based on concentrations of three metabolites (2,4-dihydroxybutanoic acid, unidentified carboxylic acid, phosphatidylcholine (PC (16:0/16:0)) in subjects at baseline, who were diagnosed with mild cognitive impairment (MCI). (a) The characteristics of the model were determined by independent testing in one out of three of the sample across 2000 cross-validation runs. (b) Beanplots of the three metabolites included in the model. (c) Two-dimensional gas chromatography coupled to time-of-flight mass spectrometry (GC × GC–TOFMS) spectra of the two metabolites included in the model, 2,4-dihydroxybutanoic acid and an unidentified carboxylic acid. Acc=classification accuracy; AUC=area under the receiver operating characteristic (ROC) curve; OR=odds ratio.
APOE genotype alone was a poor predictor of progression from MCI to AD in comparison with the predictive metabolic biomarker (P<0.001), with AUC=0.59, 90% CI=(0.47, 0.70). Addition of APOE genotype to the metabolic signature did not significantly improve the predictive model (P=0.15), with AUC=0.75, 90% CI=(0.63, 0.85; Supplementary Figure 7).
Metabolic pathways behind progression to AD
Next, we investigated which metabolic pathways may be behind the observed metabolic profile changes found to be associated with AD and with progression to AD. We applied the pathway analysis of GC × GC–TOFMS data using MPEA,38 aiming to identify sets of metabolites belonging to specific metabolic pathways, which are significantly different between (1) controls and AD groups at baseline (Figure 1 and Supplementary Figure S1) or (2) S-MCI and P-MCI groups at baseline (Figure 2). The results are shown in Table 3. The only significantly altered pathway following the P-value correction was pentose phosphate pathway when comparing P-MCI and S-MCI groups. Of relevance to this pathway, concentration of ribose-5-phosphate was decreased in the P-MCI group (P=0.046), whereas lactic acid (P=0.040) and pyruvic acid (P=0.058) were increased.
Discussion
Our findings, based on a well-phenotyped population, associate specific metabolic abnormalities with progression to AD. Our non-targeted methods cover a representative part of the main metabolic pathways, thus allowing the determination of main intermediates of lipid metabolism, energy metabolism (tricarboxylic acid cycle, gluconeogenesis, ketogenesis) and nitrogen metabolism.
At the baseline, patients diagnosed with AD had decreased concentrations of several lipid classes, including PC, plasmalogens, sphingomyelins and sterols. Plasmalogens are ether phospholipids, which are enriched in polyunsaturated fatty acids, and are abundant in the brain.41, 42 They have been found diminished in AD in multiple previous studies,43, 44, 45 as well as in normal aging.9 Also diminishment of sphingomyelins and sterols is in line with earlier findings implicating altered sterol and sphingomyelin metabolism in AD.46, 47, 48 Recent study suggests that ɛ4 allele of APOE (APOE4), a major risk allele of AD,49 is associated with disruption of sterol and sphingolipid metabolism.50 Given the affected lipids are major constituents of lipid membranes, their compositional variation with age and in disease is likely affecting the membrane fluidity and protein mobility.41, 51, 52 This is particularly relevant given recent evidence that truncated amyloid β fragments may dynamically form ion channels and may so affect the uptake of ions such as calcium into the cells.53 The membrane lipid milieu may thus be an important contributing factor modulating the dynamics of Aβ self-assembly.54
Plasmalogens via the vinyl-ether bond also act as endogenous antioxidants to protect cells from reactive oxygen species, and their diminishment in AD is in line with the hypothesis implicating the role of oxidative stress in AD pathogenesis.55, 56 In agreement with earlier studies, circulating histamine was elevated in patients diagnosed with AD.57, 58 Histamine stimulates production of nitric oxide59 and thus, the activation of the histaminergic system may also contribute to the pathology of AD.60
The metabolite biomarker signature was identified, which was predictive of progression to AD (Figure 2). The major contributing metabolite in the marker panel separating P-MCI and S-MCI patients was 2,4-dihydroxybutanoic acid. Interestingly, this organic acid is a major component of CSF,61, 62 but is found in plasma at nearly two orders of magnitude lower concentrations as in CSF.61 Very scarce data is available on the biochemistry of 2,4-dihydroxybutanoic acid. In one report, this metabolite was overproduced under low oxygen conditions from D-galacturonic acid,63 a uronic acid, which is a stereoisomer of glucoronic acid. Concentration of glucoronic acid was decreased at a marginal significance level in the P-MCI group in our study (P=0.10). In support of this interpretation, there were significant differences in the pentose phosphate pathway as shown by pathway analysis, including decrease of ribose-5-phosphate and increase of lactic acid, an end product of glycolysis. It is known that under hypoxic conditions in the brain, more glucose is metabolized via the pentose phosphate pathway.64 Studies in APP23 transgenic mice have in fact shown that hypoxia facilitates progression to AD.65
The study setting with a prospective cohort of carefully characterized and followed-up subjects is a definitive strength of the present study. This allowed us to identify the patients diagnosed with MCI, who later progressed to AD, and in deriving the molecular signature, which can identify such patients at baseline. In a health care setting, application of such a biochemical assay could therefore complement the neurocognitive assessment by the medical doctor and could be applied to identify the at-risk patients in need of further comprehensive follow-up. As a potential limitation of our study, the relatively small sample size did not allow us to split our sample into two independent cohorts. As an alternative, we performed an implicit validation by performing a model selection over a large number of randomly selected subsets of samples, then each time, independently validating the model in the rest of the sample. The most commonly selected model was then selected as our metabolic signature. This approach allowed us to estimate and report the distribution of model performances and not only of the most optimistic model, therefore providing a reasonable estimate of how the model may perform in the independent validation setting.
In conclusion, we have identified metabolic profile changes of potential pathogenic relevance in progression to and overt AD. Our findings primarily implicate the roles of hypoxia, oxidative stress, as well as membrane lipid remodeling in AD. Given the key metabolite from the metabolic signature predictive of progression to AD is abundant in CSF, further investigations should, in addition to its validation in other cohort studies, also include metabolomic studies in CSF, as well as in experimental models. Establishment of pathogenic relevance of predictive biomarkers such as ours may not only facilitate early diagnosis, but may also help identify new therapeutic avenues.
References
Qiu C, De Ronchi D, Fratiglioni L . The epidemiology of the dementias: an update. Curr Opin Psychiatry 2007; 20: 380–385.
Petersen RC . Mild cognitive impairment as a diagnostic entity. J Intern Med 2004; 256: 183–194.
Petersen RC, Stevens JC, Ganguli M, Tangalos EG, Cummings JL, DeKosky ST . Practice parameter: early detection of dementia: mild cognitive impairment (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology 2001; 56: 1133–1142.
Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K et al. Mild cognitive impairment. Lancet 2006; 367: 1262–1270.
Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med 2007; 13: 1359–1362.
O’Bryant SE, Xiao G, Barber R, Reisch J, Doody R, Fairchild T et al. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol 2010; 67: 1077–1081.
Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet 2010; 42: 137–141.
Lenz EM, Bright J, Wilson ID, Hughes A, Morrisson J, Lindberg H et al. Metabonomics, dietary influences and cultural differences: a 1H NMR-based study of urine samples obtained from healthy British and Swedish subjects. J Pharm Biomed Anal 2004; 36: 841–849.
Maeba R, Maeda T, Kinoshita M, Takao K, Takenaka H, Kusano J et al. Plasmalogens in human serum positively correlate with high-density lipoprotein and decrease with aging. J Atheroscler Thromb 2007; 14: 12–18.
Nikkilä J, Sysi-Aho M, Ermolov A, Seppänen-Laakso T, Simell O, Kaski S et al. Gender dependent progression of systemic metabolic states in early childhood. Mol Syst Biol 2008; 4: e197.
Oresic M, Simell S, Sysi-Aho M, Näntö-Salonen K, Seppänen-Laakso T, Parikka V et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med 2008; 205: 2975–2984.
Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu PV, Yetukuri L, Islam S et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res 2010; 51: 1101–1112.
Oresic M . Metabolomics, a novel tool for studies of nutrition, metabolism and lipid dysfunction. Nutr Metab Cardiovasc Dis 2009; 19: 816–824.
van der Greef J, Stroobant P, Heijden Rvd . The role of analytical sciences in medical systems biology. Curr Opin Chem Biol 2004; 8: 559–565.
Hartmann T, Kuchenbecker J, Grimm MOW . Alzheimer's disease: the lipid connection. J Neurochem 2007; 103: 159–170.
Oresic M, Hänninen VA, Vidal-Puig A . Lipidomics: a new window to biomedical frontiers. Trends Biotechnol 2008; 26: 647–652.
Han X, Gross RW . Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 2005; 24: 367–412.
Lagarde M, Geloen A, Record M, Vance D, Spener F . Lipidomics is emerging. Biochim Biophys Acta 2003; 1634: 61.
Kivipelto M, Helkala EL, Hänninen T, Laakso MP, Hallikainen M, Alhainen K et al. Midlife vascular risk factors and late-life mild cognitive impairment: A population-based study. Neurology 2001; 56: 1683–1689.
Hänninen T, Hallikainen M, Tuomainen S, Vanhanen M, Soininen H . Prevalence of mild cognitive impairment: a population-based study in elderly subjects. Acta Neurol Scand 2002; 106: 148–154.
Pennanen C, Kivipelto M, Tuomainen S, Hartikainen P, Hanninen T, Laakso MP et al. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol Aging 2004; 25: 303–310.
Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN et al. Apolipoprotein E status as a predictor of the development of Alzheimer's disease in memory-impaired individuals. JAMA 1995; 273: 1274–1278.
Smith GE, Petersen RC, Parisi JE, Ivnik RJ, Kokmen E, Tangalos EG et al. Definition, course, and outcome of mild cognitive impairment. Aging Neuropsychol Cogn 1996; 3: 141–147.
Berg L . Clinical Dementia Rating (CDR). Psychopharmacol Bull 1988; 24: 637–639.
American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th edn. American Psychiatric Association, 1994.
McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM . Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984; 34: 939–944.
Julkunen V, Niskanen E, Muehlboeck S, Pihlajamaki M, Kononen M, Hallikainen M et al. Cortical thickness analysis to detect progressive mild cognitive impairment: a reference to Alzheimer's disease. Dement Geriatr Cogn Disord 2009; 28: 404–412.
Tsukamoto K, Watanabe T, Matsushima T, Kinoshita M, Kato H, Hashimoto Y et al. Determination by PCR-RFLP of apo E genotype in a Japanese population. J Lab Clin Med 1993; 121: 598–602.
Nygren H, Seppanen-Laakso T, Castillo S, Hyotylainen T, Oresic M . Liquid Chromatography-Mass Spectrometry (LC-MS)-Based Lipidomics for Studies of Body Fluids and Tissues. Methods Mol Biol 2011; 708: 247–257.
Castillo S, Mattila I, Miettinen J, Orešič M, Hyötyläinen T . Data analysis tool for comprehensive two-dimensional gas chromatography-time of flight mass spectrometry. Anal Chem 2011; 83: 3058–3067.
Pluskal T, Castillo S, Villar-Briones A, Oresic M . MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010; 11: 395.
Kampstra P . Beanplot: a boxplot alternative for visual comparison of distributions. J Stat Soft 2008; 28 (Code Snippet 1): 1–9.
Fraley C, Raftery AE . Model-based methods of classification: Using the mclust software in chemometrics. J Stat Soft 2007; 18: 1–13.
Dalgaard P . Introductory Statistics with R. Springer Verlag: New York, 2004.
Friedman J, Hastie T, Tibshirani R . Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1–22.
Yamashita T, Yamashita K, Kamimura R . A stepwise AIC method for variable selection in linear regression. Commun Stat Theory Methods 2007; 36: 2395–2403.
McCullogh P, Nelder JA . Generalized linear models. Chapman & Hall/CRC: New York, 1989.
Kankainen M, Gopalacharyulu P, Holm L, Orešič M . MPEA—Metabolite Pathway Enrichment Analysis. Bioinformatics 2011; 27: 1878–1879.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA 2005; 102: 15545–15550.
Steuer R, Kurths J, Fiehn O, Weckwerth W . Observing and interpreting correlations in metabolomic networks. Bioinformatics 2003; 19: 1019–1026.
Brites P, Waterham HR, Wanders RJA . Functions and biosynthesis of plasmalogens in health and disease. Biochim Biophys Acta 2004; 1636: 219–231.
Farooqu AA, Horrocks LA . Plasmalogens, phospholipase A2, and docosahexaenoic acid turnover in brain tissue. J Mol Neurosci 2001; 16: 263–272; discussion 279–284.
Goodenowe DB, Cook LL, Liu J, Lu Y, Jayasinghe DA, Ahiahonu PW et al. Peripheral ethanolamine plasmalogen deficiency: a logical causative factor in Alzheimer's disease and dementia. J Lipid Res 2007; 48: 2485–2498.
Han X, Holtzman DM, McKeel Jr DW . Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem 2001; 77: 1168–1180.
Farooqui AA, Rapoport SI, Horrocks LA . Membrane phospholipid alterations in Alzheimer's disease: deficiency of ethanolamine plasmalogens. Neurochem Res 1997; 22: 523–527.
He X, Huang Y, Li B, Gong CX, Schuchman EH . Deregulation of sphingolipid metabolism in Alzheimer's disease. Neurobiol Aging 2010; 31: 398–408.
Puglielli L, Tanzi RE, Kovacs DM . Alzheimer's disease: the cholesterol connection. Nat Neurosci 2003; 6: 345–351.
Han X, Rozen S, Boyle SH, Hellegers C, Cheng H, Burke JR et al. Metabolomics in early Alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE 2011; 6: e21643.
Laws SM, Hone E, Gandy S, Martins RN . Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem 2003; 84: 1215–1236.
Bandaru VV, Troncoso J, Wheeler D, Pletnikova O, Wang J, Conant K et al. ApoE4 disrupts sterol and sphingolipid metabolism in Alzheimer's but not normal brain. Neurobiol Aging 2009; 30: 591–599.
Nagan N, Zoeller RA . Plasmalogens: biosynthesis and functions. Prog Lipid Res 2001; 40: 199–229.
Niemela PS, Hyvonen MT, Vattulainen I . Atom-scale molecular interactions in lipid raft mixtures. Biochim Biophys Acta 2009; 1788: 122–135.
Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL et al. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer's Disease and Down syndrome. Proc Natl Acad Sci USA 2010; 107: 6538–6543.
Orešič M, Lötjönen J, Soininen H . Systems medicine and the integration of bioinformatic tools for diagnosis of Alzheimer's disease. Genome Med 2010; 2: e83.
Markesbery WR . Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med 1997; 23: 134–147.
Pratico D . Oxidative stress hypothesis in Alzheimer's disease: a reappraisal. Trends Pharmacol Sci 2008; 29: 609–615.
Alvarez XA, Franco A, Fernandez-Novoa L, Cacabelos R . Blood levels of histamine, IL-1 beta, and TNF-alpha in patients with mild to moderate Alzheimer disease. Mol Chem Neuropathol 1996; 29: 237–252.
Cacabelos R, Fernandez-Novoa L, Perez-Trullen JM, Franco-Maside A, Alvarez XA . Serum histamine in Alzheimer's disease and multi-infarct dementia. Methods Find Exp Clin Pharmacol 1992; 14: 711–715.
Lantoine F, Iouzalen L, Devynck MA, Millanvoye-Van Brussel E, David-Dufilho M . Nitric oxide production in human endothelial cells stimulated by histamine requires Ca2+ influx. Biochem J 1998; 330 (Part 2): 695–699.
Haas H, Panula P . The role of histamine and the tuberomamillary nucleus in the nervous system. Nat Rev Neurosci 2003; 4: 121–130.
Hoffmann GF, Meier-Augenstein W, Stockler S, Surtees R, Rating D, Nyhan WL . Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis 1993; 16: 648–669.
Stoop MP, Coulier L, Rosenling T, Shi S, Smolinska AM, Buydens L et al. Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol Cell Proteomics 2010; 9: 2063–2075.
Niemelä K, Sjöström E . Non-oxidative and oxidative degradation of D-galacturonic acid with alkali. Carbohydrate Res 1985; 144: 93–99.
Hakim AM, Moss G, Gollomp SM . The effect of hypoxia on the pentose phosphate pathway in brain. J Neurochem 1976; 26: 683–688.
Sun X, He G, Qing H, Zhou W, Dobie F, Cai F et al. Hypoxia facilitates Alzheimer's disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci USA 2006; 103: 18727–18732.
Acknowledgements
This work was funded under the 7th Framework Programme by the European Commission (EU-Grant-224328-PredictAD; Name: From Patient Data to Personalised Healthcare in Alzheimer's Disease), Health Research Council of The Academy of Finland, Grant 121038, and EVO Grant 5772709 from Kuopio University Hospital. We thank Ulla Lahtinen, Anna-Liisa Ruskeepää, Matti Kankainen, Mika Hilvo and Sandra Castillo for their help in metabolomics analysis and data processing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Orešič, M., Hyötyläinen, T., Herukka, SK. et al. Metabolome in progression to Alzheimer's disease. Transl Psychiatry 1, e57 (2011). https://doi.org/10.1038/tp.2011.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2011.55
Keywords
This article is cited by
-
Plasma metabolomic profiles of dementia: a prospective study of 110,655 participants in the UK Biobank
BMC Medicine (2022)
-
Circulating serum metabolites as predictors of dementia: a machine learning approach in a 21-year follow-up of the Whitehall II cohort study
BMC Medicine (2022)
-
Neurodegenerative Disorders of Alzheimer, Parkinsonism, Amyotrophic Lateral Sclerosis and Multiple Sclerosis: An Early Diagnostic Approach for Precision Treatment
Metabolic Brain Disease (2022)
-
Looking at time dependent differentiation of mesenchymal stem cells by culture media using MALDI-TOF-MS
Cell and Tissue Banking (2022)
-
Huanglian Jiedu decoction remodels the periphery microenvironment to inhibit Alzheimer’s disease progression based on the “brain-gut” axis through multiple integrated omics
Alzheimer's Research & Therapy (2021)