Abstract
In yeast, the pol3-01,L612M double mutant allele, which causes defects in DNA polymerase delta (Pol δ) proofreading (pol3-01) and nucleotide selectivity (pol3-L612M), confers an “ultramutator” phenotype that rapidly drives extinction of haploid and diploid MMR-proficient cells. Here, we investigate antimutator mutations that encode amino acid substitutions in Pol δ that suppress this lethal phenotype. We find that most of the antimutator mutations individually suppress the pol3-01 and pol3-L612M mutator phenotypes. The locations of many of the amino acid substitutions in Pol δ resemble those of previously identified antimutator substitutions; however, two novel mutations encode substitutions (R674G and Q697R) of amino acids in the fingers domain that coordinate the incoming dNTP. These mutations are lethal without pol3-L612M and markedly change the mutation spectra produced by the pol3-01,L612M mutator allele, suggesting that they alter nucleotide selection to offset the pol3-L612M mutator phenotype. Consistent with this hypothesis, mutations and drug treatments that perturb dNTP pool levels disproportionately influence the viability of pol3-L612M,R674G and pol3-L612M,Q697R cells. Taken together, our findings suggest that mutation rate can evolve through genetic changes that alter the balance of dNTP binding and dissociation from DNA polymerases.
Similar content being viewed by others
Introduction
Cells must accurately duplicate their genome each division to ensure that their descendants inherit the proven genetic traits of past generations. The greatest contribution to high fidelity DNA synthesis comes from the inherent nucleotide selectivity of the main replicative DNA polymerases, which make one misinsertion every 105 to 106 base-pairs. Extension of the resulting mispaired primer termini by replicative polymerases normally occurs inefficiently1,2,3,4. Pausing provides time for mismatched primer termini to partition to an intrinsic or extrinsic 3′ → 5′ proofreading exonuclease5,6,7, which removes the errant 3′ nucleotide. If polymerases extend the mismatch before proofreading occurs, post-replicative mismatch repair (MMR) complexes recognize the distorted duplex DNA and initiate excision and re-synthesis of the nascent strand8,9,10,11,12. Together, nucleotide selectivity, proofreading, and MMR lower the DNA replication error rate to less than 1 × 10−9 mutations per base pair per cell division13.
In eukaryotes, DNA polymerases epsilon (Pol ε) and delta (Pol δ) are thought to perform the bulk of leading and lagging strand DNA synthesis, respectively14,15,16. Proofreading-deficient Pol ε and Pol δ variants elevate mutation rates in yeast8,17,18,19 and mouse cells20,21,22. Consistent with the hypothesis that high DNA replication fidelity restrains neoplasia, Pol ε and Pol δ proofreading defects increase cancer risk in mice20,21,22 and recently have been found in highly mutated human colorectal and endometrial cancers23,24,25,26,27,28, complementing studies from the 1990’s linking the Lynch cancer susceptibility syndrome to MMR defects29.
Since nucleotide selectivity, proofreading and MMR act redundantly to limit the transmission of DNA polymerase errors, combined defects in any two of these activities lead to strong synergistic mutator phenotypes in diploid yeast, with mutation rates 1000–50,000 fold more than that of wild-type cells8,19,30,31,32. Recent evidence suggests that such “ultramutator” cells may emerge spontaneously in certain human cancers. The most dramatic demonstration comes from patients with bi-allelic MMR deficiencies who spontaneously developed brain tumors with additional mutations affecting nucleotide selectivity or proofreading33.
The emergence of cell lineages with synergistic mutator alleles conforms to evolutionary theory. Mutator alleles hitch-hike on the fitness benefits of the adaptive mutations they generate34,35,36,37,38, and an additional increase in mutation rate fuels even more rapid adaptation. But the accumulation of deleterious mutations in mutator cells takes a toll. In bacteria and both haploid and diploid yeast, mutator phenotypes that exceed an error threshold drive extinction of the mutator lineage within a few cell divisions due to inactivation of essential genes, a process we refer to as Error-induced Extinction (EEX)32,39,40,41,42,43. Evidence also exists for error thresholds in multicellular organisms. Mice that are homozygous for mutant alleles that cause defects in MMR and either Pol δ or Pol ε proofreading are embryonic lethal22.
Intriguingly, human tumor cells with tandem deficiencies in MMR and proofreading do not appear to exceed a mutation load of ~250 × 10−6 mutations/bp25,33. Diploid yeast cells with mutation rates near the threshold of error-induced extinction carry similar mutation burdens of ~70 × 10−6 mutations/bp and exhibit highly variable colony forming capacity32. Thus, the apparent upper limit of mutation load in ultramutator tumors may derive from an attrition of the most highly mutated cells, whose probability of getting a lethal mutation in the next round of replication increases proportionately with the number of recessive lethal mutations they carry.
Yeast cells undergoing EEX experience strong selection pressure for eex mutants with “antimutator” alleles that lower mutation rates to tolerable levels38,39,44,45,46,47. In this present study, we investigate eex mutations that suppress the ultramutator phenotype of a double mutant allele (pol3-01,L612M) created by combining the classic pol3-01 mutation, which compromises proofreading, with a mutation encoding an L612 to M substitution in the Pol δ active site32,48. L612 is highly conserved and forms part of the binding site for the incoming dNTP49. Early studies by Reha-Krantz and colleagues with Phage T4 polymerase established that the equivalent L412M substitution conferred a mutator phenotype50. The corresponding substitution in yeast Pol δ increases mutation rate modestly, producing primarily base-substitution errors48,51. The same change in human Pol δ (L606M) increases the error rate in vitro52 and was recently observed in a hyper-mutated brain tumor from one of the above patients with bi-allelic MMR deficiency33. We previously showed that the pol3-01,L612M allele induces both haploid and diploid yeast cells to undergo error-induced extinction32, with the inferred mutation rate of pol3-01,L612M diploids being ~50,000 times greater than that of wild-type cells32. We found that the eex mutations that suppressed pol3-01,L612M lethality exhibited a wide range of antimutator effects (reprinted in Table 1)32. In theory, the eex alleles could specifically suppress the pol3-01 or pol3-L612M mutator alleles or exert a general antimutator effect on Pol δ fidelity. Here, we explore this question of allele-specificity and examine the structural implications of the eex substitutions in order to understand their underlying antimutator mechanisms.
Results
Interactions between mutator and antimutator alleles
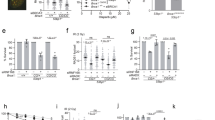
To test the allele-specificity of the antimutator alleles, we generated pol3-01,eex and pol3-L612M,eex double mutant alleles, as well as pol3-eex single mutant alleles, and then examined their influence on viability and mutation rates in the presence and absence of MMR. Mutations conferring MMR-deficiency are synthetically lethal with pol3-01 in haploids, but not in diploids. Thus, we utilized wild-type and Msh2-deficient diploid “plasmid shuffling” strains, which were engineered to be hemizygous for CAN1 to permit measurement of forward mutation rates32. The strains carry deletions of both chromosomal POL3 genes, complemented by a POL3-URA3-CEN plasmid. We transformed these cells with wild-type and mutant POL3 alleles carried on LEU2-CEN plasmids, and then assessed their influence on viability by plating each strain-plasmid combination on FOA plates, which selects for loss of the POL3-URA3 plasmid (~1 in 100 cells) (Fig. 1A). For viable strains, the FOA-resistant colonies were then used as replica colonies for mutation rate determination by fluctuation analysis53,54.
(A) Top. Plasmid shuffling scheme. Diploid plasmid shuffling strains with chromosomal deletions of POL3 (lines with triangles) complemented by a POL3–URA3 plasmid (blue) were transformed with LEU2 plasmids (red) expressing various POL3 alleles (pol3-LEU2) on plates lacking leucine and uracil (-Leu -Ura). Bottom. The above pol3-LEU2 transformants were then plated on FOA media in 10-fold serial dilutions to assess viability. FOA-resistant colonies were used as replica cultures in a fluctuation assay to determine mutation rates conferred by (B) pol3-eex alleles, (C) pol3-01,eex, (D) pol3-L612M,eex and (E) pol3-01,L612M,eex (reprinted from Herr et al.32). Black bars indicate MMR proficient cells (BP8001). White bars indicate msh2Δ/msh2Δ cells (BP9101). “X” indicates no growth.
MMR-proficient diploids grew with most of the mutant eex alleles as the sole source of Pol δ, indicating that the corresponding amino acid substitutions do not substantially impair Pol δ activity (Fig. 1A). The exceptions were the pol3-eex and pol3-01,eex alleles with R674G or Q697R. The lethality of these alleles suggests that the R674G and Q697R substitutions inhibit Pol δ function in the absence of L612M. Three of the pol3-eex alleles (S319F, F467I, and K559N) increased mutation rate significantly in the presence of MMR by a likelihood ratio test, recently developed by Zheng54 (Fig. 1B, Table 1). In the absence of Msh2, four alleles (pol3-S319F, pol3-R658G, pol3-A704V, and pol3-G818C) increased mutation rates significantly, suggesting that the corresponding polymerases create errors that MMR corrects. In contrast, the absence of a synergistic effect between pol3-F467I and msh2Δ, suggests that the errors triggered by Pol δ-F467I are not subject to MMR. Unexpectedly, the pol3-K559N allele conferred poor colony forming capacity in the absence of MMR (Fig. 1A), which precluded measuring mutation rates. Proofreading deficiency may exacerbate this synthetic phenotype, as the pol3-01,K559N allele was synthetically lethal with msh2Δ (Fig. 1A).
Although some of the eex alleles confer mutator phenotypes on their own, they exert clear antimutator effects on pol3-01 and pol3-L612M (Fig. 1C,D). All non-lethal eex alleles suppressed the pol3-01 mutator phenotype to levels observed with previously isolated pol3-01,eex alleles39. The eex alleles exerted more modest antimutator effects on pol3-L612M (Table 1), which were more apparent in msh2Δ cells, where the majority lowered mutation rates more than 3-fold (Table 1). Thus, although the eex alleles could theoretically have been specific to pol3-01 or pol3-L612M, most appear to increase fidelity by perturbing an aspect of Pol δ activity required for both mutator mechanisms.
Influence of antimutator alleles on pol3-01,L612M mutation spectra
In order to understand how the eex mutations altered the pol3-01,L612M mutator phenotype, we performed whole genome sequencing of four Msh2-deficient pol3-01,L612M,eex strains that express Pol δ variants with amino acid substitutions near the polymerase active site (S611Y, R674G, Q697R, and K559N). The canavanine-resistance mutation rate exhibited by each of these strains is at least 40 times higher than the POL3 Msh2-deficient control (Fig. 1E, Table 1); thus, most mutations in these strains should derive from errors caused by the mutant Pol δ variants. We obtained independent isolates of these strains by plasmid shuffling. After they formed a colony of 105 to 106 cells, which corresponds to 17–20 generations of growth, we isolated single cells from the original colonies and allowed them to form new colonies. We then identified the accumulated mutations by whole genome sequencing, scoring both the frequency of different classes of mutations as well as their relative fraction of the total mutations observed. As controls, we sequenced similarly isolated MMR-deficient strains expressing pol3-01, pol3-L612M, or both POL3 and pol3-01,L612M (POL3/pol3-01,L612M). We relied on these “episomally heterozygous” POL3/pol3-01,L612M cells to define the spectrum of mutations generated by Pol δ-01,L612M, since cells expressing only the pol3-01,L612M allele are inviable.
The MMR-deficient POL3/pol3-01,L612M isolates averaged more than 1700 mutations per genome (Table S1). If we assume that these mutations occurred over ~20 generations, the Pol δ-01,L612M polymerase caused a genome-wide mutation rate of 89 mutations per cell division (Table S1). The pol3-01 strains also averaged more than 1700 mutations per genome, while pol3-L612M strains averaged only 566 mutations per genome, consistent with a lower mutation rate (Table S1). Thus, although heterozygous, the pol3-01,L612M allele generates sufficient numbers of mutations for meaningful comparisons with the mutation spectra from other strains.
As observed previously in a POL3/pol3-01,L612M MMR-proficient diploid32, the most abundant mutations observed in the POL3/pol3-01,L612M MMR-deficient cells were C → T/G → A transitions, followed by A → G/T → C transitions and C → A/G → T transversions, which were 50% less frequent than C → T/G → A transitions (Fig. 2, Table S1). Like POL3/pol3-01,L612M cells, cells with pol3-01 or pol3-L612M generated roughly half as many A → G/T → C transitions as C → T/G → A transitions; however, both had a lower fraction of C → A/G → T transversions than the POL3/pol3-01,L612M cells (Fig. 2B). The MMR-deficient pol3-01,L612M,S611Y cells, which averaged more than 2400 mutations per genome, exhibited a mutation spectrum similar to pol3-01 cells (Fig. 2B). Cells expressing pol3-01,L612M,eex alleles with the K559N, R674G, or Q697R mutations had overall mutation frequencies that were similar to pol3-L612M cells (Fig. 2A). Like pol3-L612M cells, they produced a smaller fraction of C → A/G → T transversion mutations than POL3/pol3-01,L612M cells. However, each pol3-01,L612M,eex strain produced proportionally similar levels of C → T/G → A and A → G/T → C transitions, in contrast to the POL3/pol3-01,L612M, pol3-01, and pol3-L612M strains (Fig. 2B, Table S1). The striking similarity in mutation spectra induced by these eex alleles suggests that they suppress the ultramutator phenotype of pol3-01,L612M by a common mechanism that influences nucleotide selectivity.
We clonally isolated cells expressing mutator alleles after 17–20 generations of growth and performed whole genome sequencing. (A) Mutation frequencies. The frequencies of transitions and transversions were determined by dividing the number of observations of each class by the number of scored sites in the genome that could give rise to that mutation. The frequency of each class of insertion/deletion mutation (indels) were determined by dividing the number of observations by the total number of scored sites. (B) Mutation Fractions. The fraction of each class of mutation was determined by dividing the number of observation of each class by the total number of observed mutations. The error bars for both (A) and (B) represent the standard deviations of the mutation frequencies and fractions obtained from independent isolates of the same genotype.
Mutations and drug treatments known to perturb dNTP pool levels influence the viability of cells expressing pol3-L612M,R674G or pol3-L612M,Q697R
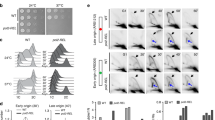
Nucleotide selectivity is influenced by the composition and levels of dNTP pools, which are regulated in yeast by the Damage uninducible (Dun) 1 kinase as part of the S-phase checkpoint55 and during normal dNTP homeostasis56,57. Elimination of Dun1 (dun1Δ) suppresses the lethality of the pol3-01,L612M ultramutator allele and the mutator phenotypes of most pol3-01,L612M,eex alleles32, as well as pol3-0158, pol2-457, and pol3-R696W59. Intriguingly, pol3-01,L612M,R674G and pol3-01,L612M,Q697R are synthetically lethal with dun1Δ32, suggesting that the R674G and Q697R substitutions may lower dNTP binding affinity of Pol δ, thereby imposing a requirement for higher dNTP pools. Dun1 increases dNTP pools by upregulating ribonucleotide reductase (RNR) — the rate limiting enzyme for dNTP synthesis — in part, by targeting Suppressor of mec1 lethality (Sml)160, which interferes with RNR complex assembly (Fig. 3A)61. Previous work has shown that while dun1Δ mutant cells have a two-fold reduction in dNTP pools relative to wild-type cells, dun1Δ sml1Δ mutant cells have a 2-fold increase in dNTP pools relative to wild-type cells62.
(A) Regulation of dNTP synthesis. Phosphorylated Dun1 increases dNTP synthesis by negatively regulating Dif1 (Damage-regulated Import facilitator 1)78 Crt1, (Constitutive RNR Transcription 1)79, and Sml1. These three proteins repress either RNR gene expression or RNR assembly. (B) Genetic interactions with dun1Δ and sml1Δ. Two independent plasmid shuffling strains per genotype were plated onto FOA media in ten-fold serial dilutions. (C) Sensitivity to HU. Two independently shuffled strains per genotype were plated in ten-fold serial dilutions onto media containing increasing concentrations of HU.
To test the hypothesis that insufficient dNTP pools drives the synthetic lethality between dun1Δ and the R674G or Q697R mutations, we assessed the viability of dun1Δ and dun1Δ sml1Δ haploid strains expressing the pol3-L612M,R674G or pol3-L612M,Q697R alleles. While Dun1 deficiency impaired growth of cells expressing either allele, Sml1 deficiency ameliorated this effect (Fig. 3B). As a second test of the hypothesis, we assessed the sensitivity of DUN1-proficient pol3-L612M,R674G or pol3-L612M,Q697R mutants to increasing concentrations of hydroxyurea (HU), which blocks dNTP synthesis by inhibiting RNR activity (Fig. 3A). We found that both pol3-L612M,eex alleles impaired colony growth at concentrations of HU that were tolerated by cells expressing POL3 or pol3-L612M (Fig. 3C). These observations support the hypothesis that R674G and Q697R enhance replication fidelity by reducing the dNTP binding affinity of Pol δ.
Discussion
The evolution of mutation rate occurs through the acquisition of mutator and antimutator traits that optimize DNA replication fidelity63. The most radical antimutator innovations of evolution are unquestionably proofreading and MMR, which involved the recruitment of protein domains or entire complexes to enhance the accuracy of DNA replication. But most mutator and antimutator alleles likely encode single amino acid substitutions in proteins already involved in DNA replication. The mechanism by which a mutator allele elevates mutation rate may determine what antimutator mutations restore fidelity. Both general and allele-specific antimutators may be possible. We analyzed antimutator alleles that suppress a synergistic mutator phenotype caused by two different mutator alleles affecting Pol δ nucleotide selectivity and proofreading. We found that most antimutator mutations exert a suppressive effect on each mutator allele individually, suggesting that they limit an aspect of Pol δ activity required for all mutagenesis, such as the extension of mispaired primer termini. However, we did find allele-specific interactions between pol3-L612M and the R674G and Q697R mutations, which influenced cellular viability. In what follows, we discuss the basis for the ultramutator phenotype of pol3-01,L612M and how the amino acid substitutions encoded by the antimutator mutations may influence replication fidelity and allele-specific viability in light of their locations within the Pol δ structure.
Kinetic experiments in the late 1980’s revealed that high-fidelity DNA polymerases extend mispaired primer termini inefficiently, which augments proofreading1,2,3,4,64. In the absence of proofreading, Pol δ can eventually extend errors into duplex DNA65. Biochemical studies indicate that the L612M substitution lowers this kinetic barrier, allowing Pol δ to frequently extend mispairs even with an intact proofreading domain66. If failure to proofread mispaired primer termini was the only source of Pol δ-L612M infidelity, the mutator phenotype of pol3-01 would be epistatic to that of pol3-L612M. Instead, the robust pol3-01,L612M mutator phenotype far exceeds that of the individual pol3-01 or pol3-L612M alleles32. Thus, in addition to promoting mispair extension, the L612M substitution must also decrease nucleotide selectivity, which is supported by the distinctive pol3-L612M error signature14 and the increase in C → A/G → T transversions in the POL3/pol3-01,L612M strain. Of course, increased mispair extension and lowered nucleotide selectivity may derive from the same phenomenon, such as increased dNTP binding stability or higher catalytic activity.
Pol δ resembles a right hand with five domains including the amino-terminal domain (amino), the exonuclease domain (exo), the palm (containing the catalytic metal ions for DNA synthesis), the fingers, and the thumb (Fig. 4A)49. Many of the antimutator substitutions are relatively far from the polymerase active site, near previously identified substitutions encoded by mutations that suppress the pol3-01 mutator phenotype39. The R658G substitution alters an interaction between the palm and the amino domains, also affected by the antimutator substitutions G204D, G207R, E642K, and D643N (Fig. 4B)39,67. S319F resides in the central β-sheet located in the heart of the exo domain, adjacent to the G400S antimutator substitution (Fig. 4C)39. The F467I substitution affects a helix-loop-helix motif in the exo domain that interacts with the backbone of the primer strand and contains the L531P and F486S antimutator substitutions (Fig. 4C)39. The third substitution in the exo domain, G447D, alters a β-hairpin that contacts the DNA during polymerization (Fig. 4C,D)68,69. Consistent with a role in DNA binding stability, Reha-Krantz and colleagues previously found that a different substitution at this position, G447S, suppresses frameshift mutations, while G447D increases the rate of -1 frameshift mutations70. Finally, the G818C substitution maps to the palm domain in a different β-hairpin affected by the Y808C and W821C antimutator substitutions. This β-hairpin contains the highly conserved KKR motif (Figure S1), which binds the minor groove of the primer•template at the −3 to −5 positions and may allow Pol δ to sense deviations from Watson-Crick base-pairing geometry in the newly synthesized DNA and initiate proofreading even after mispair extension7,49 (Fig. 4E).
The S. cerevisiae Pol δ structure49 (Protein database accession code 3IAY) is shown as a schematic diagram of the α-carbon backbone (rendered in PyMol). Structural domains are color coded as follows: amino (gray), exo (red), palm (purple), fingers (blue) and thumb (green). Yellow spheres correspond to eex mutations from the present study and are labeled to indicate the amino acid substitution; light blue spheres correspond to antimutator mutations from a previous study39. The incoming dNTP is denoted by green CPK sticks and the template nucleotide by orange CPK sticks. The primer DNA is represented by tan sticks and the template DNA, by brown sticks. Important non-mutated residues proximal to the antimutator substitutions are shown as space-filling spheres and color coded according to the domain. Active-site carboxylate side chains are gray CPK sticks coming out of the purple (palm) or red (exo) ribbons. Metal ions are small black spheres. (A) Overall distribution of eex amino acid substitutions. The α-carbons are indicated by color coded spheres. (B) eex substitutions affecting an interaction between the amino and palm domains. (C) eex substitutions with structural roles in exonuclease domain. The last three nucleotides of the DNA primer strand from the RB69 Pol Editing structure6 (Protein database accession code 1CLQ) were placed in the Pol δ exo active site (tan sticks) by aligning the conserved exo domain motifs of RB69 Pol and Pol δ in Pymol. (D) Interaction of the β-hairpin of the exonuclease domain with the primer strand. (E) G818C and the KKR motif implicated in binding of duplex DNA in the minor groove7,49. (F) Amino acid substitutions affecting the polymerase active site. The ribose of the incoming dNTP stacks on Y613 and L612 (pink spheres). The γ-phosphate of the incoming dNTP contacts R674 and the primary amino group of S611. Q697 forms a hydrogen bond with R674, while K559N forms a hydrogen bond with S703.
The remaining antimutator substitutions from this study alter the polymerase active site, which is formed by elements of the amino, palm and fingers domains (Fig. 4F). The fingers domain is a dynamic structure that rapidly moves from an open to closed conformation upon dNTP binding. Closure of the fingers stabilizes correct dNTP•template interactions and may help the polymerase to reject incorrect nucleotides47. The A704V substitution occurs at the interface between the two fingers, adjacent to critical residues (N705-Y708) that form the binding site for the template•dNTP basepair. K559N resides in part of the amino domain previously implicated in pol3-01 suppression (Figure S1)39,47, and forms a hydrogen bond to S703 of the fingers. The remaining substitutions (S611Y, R674G, and Q697R) alter highly conserved residues involved in dNTP binding. R674 in the fingers domain directly interacts with the γ-phosphate of the incoming dNTP through its guanido group49. Q697, also in the fingers domain, positions R674 to contact the incoming dNTP, while S611 in the palm domain interacts with the dNTP γ-phosphate and the guanido group of R674 (Fig. 4F).
As we proposed previously47, antimutator substitutions that impair DNA binding, either during polymerization or during proofreading, may promote access of mispaired primer termini to other DNA repair pathways that remove the error. Conceivably, some structural perturbations may propagate to the polymerase active site in a manner that enhances nucleotide selectivity. A more compelling case for enhanced nucleotide selectivity can be made for those substitutions that directly impact the polymerase active site and alter the mutation spectrum of pol3-01,L612M mutator cells. The most dramatic of these, R674G, eliminates a key interaction between the fingers and the γ-phosphate of the incoming dNTP. The Q697R substitution may closely phenocopy this dNTP binding defect by displacing the R674 sidechain through charge repulsion between the two arginine side chains. The hypothesis that R674G and Q697R weaken Pol δ dNTP binding affinity is supported by the lethal phenotypes of the pol3-R674G and pol3-Q697R alleles, the suppression of this lethality by L612M, and the sensitivity of pol3-L612M,R674G and pol3-L612M,Q697R strains to mutations and drug treatments known to lower dNTP pool levels. Defects to dNTP binding will necessarily also impact the efficiency of mispair extension. Depending on the genetic background and growth conditions, a primer extension defect may promote the correction of Pol δ errors by alternative editing pathways (in the presence of DUN1 and L612M) or potentially induce lethal replication stress (in the absence of DUN1 or L612M or the presence of HU). Exploring the above models will require detailed biochemical studies of the mutant polymerases as well as genetic screens for genes encoding candidate alternative editing activities.
Ultramutator phenotypes are inherently unstable in yeast. It remains to be shown whether this is also true in cancer cells. The apparent mutation threshold observed in tumors deficient in proofreading and MMR33 may simply reflect the most probable mutation load of an ultramutator cancer cell at the time of malignant transformation. Importantly, the sequencing methodologies used in these studies are blind to whether individual cells within the ultramutated tumors continue to fix mutations at a high rate. Nevertheless, the existence of a mutation threshold in diploid yeast supports the hypothesis of an error threshold in human cancers. If a threshold exists, the outer limits of mutation load may be determined by a loss of replicative fitness or by the suppressive effects of immune recognition of newly arising neoantigens. Given enough cell proliferation, either mechanism may favor the emergence of antimutator subpopulations of cells within the tumor. If antimutators do arise, the underlying mechanism may create new vulnerabilities that could be exploited for therapy. For instance, antimutators that suppress the mutator phenotype by promoting dissociation of the polymerase from mispaired primer•templates may be dependent on a “targetable” DNA repair pathway to correct the errors. Likewise, antimutators that destabilize dNTP binding in the active site may confer sensitivity to drugs that dampen dNTP pool levels.
Materials and Methods
Media and Growth Conditions
Yeast were grown at 30 °C using YPD, synthetic complete (SC) media or SC ‘drop-out’ media deficient in defined amino acids to select for prototrophic genetic markers71. Premade nutrient supplements for SC and SC lacking uracil and leucine were purchased from Bufferad. Other drop-out nutrient supplements were made as described71 from individual components purchased from Sigma-Aldrich or Fisher Scientific. To select for Ura− cells during plasmid shuffling, we used SC media containing 1 mg/ml 5-fluroorotic-acid (FOA; Zymo Research)72. For mutation rate assays, we used SC plates containing 60 μg/ml canavanine and 200 μg/L geneticin (G418, Sigma-Aldrich) with mono-sodium-glutamate (MSG) (1 g/L) as the nitrogen source rather than ammonium sulfate73. To select for hygromycin resistance during strain construction (see below), we used YPD plates with 300 μg/ml hygromycin (Corning). For hydroxyurea (HU; MP Biomedicals, LLC) plates, we dissolved HU in 2xYPD at concentrations of 200 mM, 100 mM, 50 mM, and 0 mM. We filter-sterilized each solution, and then combined it with sterile 2x (4%) agar to obtain plates with 100 mM, 50 mM, 25 mM, and 0 mM HU.
Yeast Plasmids and Strains
Plasmids
pGL31017,74 is derived from YCp50 (CEN4/ARS1/URA3)75 and carries POL3 under control of the endogenous promoter. YCplac111POL3 and YCplac111pol3 derivatives39 are derived from YCplac111 (CEN6/ARS1/LEU2)76, and carry the wild-type or mutant pol3 coding and regulatory sequences, cloned between the HindIII and EcoRI restriction sites. Construction of YCplac111pol3-L612M and YCplac111pol3-01,L612M were described previously48. We utilized conventional cloning or the Quikchange protocol to re-engineer the eex mutations into YCplac111POL3 and YCplac111pol3 vectors.
Strains
The MMR proficient (BP8001) and MMR deficient (BP9101) diploid POL3 plasmid shuffling strains as well as the Dun1-proficient (BP7801) and dun1Δ (BP8101) haploid POL3 plasmid shuffling strains have been described previously32. To create a dun1Δ sml1Δ strain derivative of BP8101 (BP8101-T1), we deleted the SML1 gene by transforming the strain with the hygromycin-resistance gene (HphMX4), amplified from pFvL10077 using the previously described sml1U and sml1D primers57, which contain 50 nucleotides upstream and downstream of the SML1 coding sequence. We confirmed the deletion/insertion using flanking primers sml1-744F (5′-CGCGCTGAGCCCAAACGGGCTCCACTA-3′) and sml1-1745R (5′-GGCTCCTGTGTGACTCTATGGGAGGGAAGGA-3′), which amplify a 960 bp fragment in SML1 cells and a 2414 bp fragment in the sml1Δ::HphMX cells.
Plasmid shuffling
Plasmid shuffling with pGL310-containing strains has been described previously17,39,72. Cells transformed with YCplac111pol3 plasmids, YCplac111POL3 (positive control), or YCplac111 (empty vector control) were plated on SC lacking uracil and leucine. After two to three days incubation at 30 °C, individual colonies were picked and dispersed in sterile H2O. Serial dilutions containing approximately 105, 104, 103, and 102 cells were plated on SC or SC with FOA to select for cells that had spontaneously lost pGL310.
Mutation Rates
Mutation rates were determined by fluctuation analysis of canavanine-resistant mutants in replica cultures53 as described previously32. To measure forward mutation rates in diploids, we engineered our pol3Δ/pol3Δ deletion strains to be hemizygous for the CAN1 gene. Forward mutation rate assays using hemizygous target genes in diploid yeast are confounded by high rates of mitotic recombination, which produce phenotypically mutant colonies. Thus, these strains carry the kanMX transgene downstream of the functional CAN1 allele to allow selection against mitotic recombinants on canavanine selection plates with G418. This approach lowers the background of Canr colonies down to the level observed in CAN1 haploid cells.
Mutation rates and statistical tests were calculated using the R statistical package, rSalvador54, from the number of mutant colonies in each replica by first estimating m by maximum likelihood using newton.LD.plating and then dividing by the number of cell divisions inferred from the average number of colony forming units in the replica cultures. Confidence intervals were calculated using confint.LD.plating, which relies on likelihood ratios. Statistical comparisons between mutation rates were performed using a likelihood ratio test (LRT.LD.plating). Bonferroni corrections were made in R using p.adjust.
Mutation Spectra
To determine the mutation spectra of diploid mutator strains, we used plasmid shuffling to introduce the pol3 alleles into BP9101, selecting transformants on SC plates lacking uracil and leucine. We wanted to score the mutation spectra after 17 to 20 cellular generations to allow sufficient numbers of mutations to accumulate. Since pol3-01,L612M is lethal as the sole source of Pol δ in these strains, we maintained POL3/pol3-01,L612M cells in the heterozygous state by serially diluting transformants on SC plates lacking uracil and leucine to get well-isolated, independent colonies. For all other alleles, we first serially diluted the colonies on FOA media to isolate single colonies in which the LEU2-CEN plasmid was the sole source of Pol δ. We then subcloned these colonies on FOA media. Colonies selected for whole genome sequencing were grown overnight in 3 ml YPD, and genomic DNA was purified from 108 cells using a ZR Fungal/Bacterial Miniprep kit (Zymo Research). DNA was simultaneously fragmented and ligated to Illumina DNA adapters using the Nextera V2 Kit (Illumina), post-indexed by PCR, and sequenced using 101 bp, paired-end reads on an Illumina 2500 platform.
Reads were aligned to the S. cerevisiae S288C genome (Assembly R64-1-1) using the Burrows-Wheeler Aligner (Li-a). The aligned reads were then filtered to remove unmapped reads, non-uniquely mapping reads, and PCR duplicates using Picard and Samtools using standard settings (Li-b, URL: http://picard.sourceforge.net). After processing, the final average sequencing depth ranged between 100 and 300-fold. Coding variants were identified using VarScan2 with the strand bias filter option invoked (Kobolt). Only regions of the genome with >20-fold coverage that were not the site of a strain-specific SNP were evaluated32. Genomic positions where VarScan2 indicated a variant occurred between 40 and 60 percent of the reads mapping to that position were manually verified as heterozygous. Those that fell outside this range or occurred in low complexity regions, as determined by RepeatMasker, were not scored.
We assessed mutation frequencies by dividing the number of each class of mutations by the number of scored nucleotides in the genome that could produce that class of mutation (e.g. C → T/G → A mutations divided by all G or C bases). For frameshifts and larger insertion/deletion (indel) mutations we divided the number of mutations by the total number of bases sequenced. We determined the fraction of each mutation class by dividing the number of observations by the total mutations observed.
Additional Information
How to cite this article: Dennis, D. G. et al. Normally lethal amino acid substitutions suppress an ultramutator DNA Polymerase δ variant. Sci. Rep. 7, 46535; doi: 10.1038/srep46535 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Perrino, F. W. & Loeb, L. A. Differential extension of 3′ mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J. Biol. Chem. 264, 2898–2905 (1989).
Perrino, F. W., Preston, B. D., Sandell, L. L. & Loeb, L. A. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 86, 8343–8347 (1989).
Petruska, J. et al. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl. Acad. Sci. USA 85, 6252–6256 (1988).
Mendelman, L. V., Petruska, J. & Goodman, M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J. Biol. Chem. 265, 2338–2346 (1990).
Shevelev, I. V. & Hübscher, U. The 3′–5′ exonucleases. Nat Rev Mol Cell Biol 3, 364–376 (2002).
Shamoo, Y. & Steitz, T. A. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99, 155–166 (1999).
Franklin, M. C., Wang, J. & Steitz, T. A. Structure of the replicating complex of a pol a family DNA polymerase. Cell 105, 657–667 (2001).
Morrison, A., Johnson, A. L., Johnston, L. H. & Sugino, A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae . EMBO J. 12, 1467–1473 (1993).
Strand, M., Prolla, T. A., Liskay, R. M. & Petes, T. D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365, 274–276, doi: 10.1038/365274a0 (1993).
Prolla, T. A., Pang, Q., Alani, E., Kolodner, R. D. & Liskay, R. M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265, 1091–1093 (1994).
Johnson, R. E., Kovvali, G. K., Prakash, L. & Prakash, S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J. Biol. Chem. 271, 7285–7288 (1996).
Marsischky, G. T., Filosi, N., Kane, M. F. & Kolodner, R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10, 407–420 (1996).
Preston, B. D., Albertson, T. M. & Herr, A. J. DNA replication fidelity and cancer. Semin. Cancer Biol. 20, 281–293, doi: 10.1016/j.semcancer.2010.10.009 (2010).
Nick McElhinny, S. A., Gordenin, D. A., Stith, C. M., Burgers, P. M. J. & Kunkel, T. A. Division of labor at the eukaryotic replication fork. Mol. Cell 30, 137–144, doi: 10.1016/j.molcel.2008.02.022 (2008).
Clausen, A. R. et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol 22, 185–191, doi: 10.1038/nsmb.2957 (2015).
Yu, C. et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell 56, 551–563, doi: 10.1016/j.molcel.2014.09.017 (2014).
Simon, M., Giot, L. & Faye, G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 10, 2165–2170 (1991).
Morrison, A., Bell, J. B., Kunkel, T. A. & Sugino, A. Eukaryotic DNA polymerase amino acid sequence required for 3′ → 5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 88, 9473–9477 (1991).
Tran, H. T., Gordenin, D. A. & Resnick, M. A. The 3′ → 5′ exonucleases of DNA polymerases δ and ε and the 5′ → 3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae . Mol. Cell. Biol. 19, 2000–2007 (1999).
Goldsby, R. E. et al. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat. Med. 7, 638–639 (2001).
Goldsby, R. E. et al. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc. Natl. Acad. Sci. USA 99, 15560–15565 (2002).
Albertson, T. M. et al. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. USA 106, 17101–17104, doi: 10.1073/pnas.0907147106 (2009).
Church, D. N. et al. DNA polymerase ɛ and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet., doi: 10.1093/hmg/ddt131 (2013).
Yoshida, R. et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet. 19, 320–325, doi: 10.1038/ejhg.2010.216 (2011).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337, doi: 10.1038/nature11252 (2012).
Palles, C. et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 45, 136–144, doi: 10.1038/ng.2503 (2012).
Kandoth, C. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73, doi: 10.1038/nature12113 (2013).
Shinbrot, E. et al. Exonuclease mutations In DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res., doi: 10.1101/gr.174789.114 (2014).
Lynch, H. T. et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 76, 1–18, doi: 10.1111/j.1399-0004.2009.01230.x (2009).
Morrison, A. & Sugino, A. The 3′ → 5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae . Mol. Gen. Genet. 242, 289–296 (1994).
Greene, C. N. & Jinks-Robertson, S. Spontaneous frameshift mutations in Saccharomyces cerevisiae: accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics 159, 65–75 (2001).
Herr, A. J., Kennedy, S. R., Knowels, G. M., Schultz, E. M. & Preston, B. D. DNA replication error-induced extinction of diploid yeast. Genetics 196, 677–691, doi: 10.1534/genetics.113.160960 (2014).
Shlien, A. et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet., doi: 10.1038/ng.3202 (2015).
Chao, L. & Cox, E. C. Competition between high and low mutating strains of Escherichia coli . Evolution 37, 125–134 (1983).
Mao, E. F., Lane, L., Lee, J. & Miller, J. H. Proliferation of mutators in a cell population. J. Bacteriol. 179, 417–422 (1997).
Sniegowski, P. D., Gerrish, P. J. & Lenski, R. E. Evolution of high mutation rates in experimental populations of E. coli . Nature 387, 703–705 (1997).
Nilsson, A. I., Kugelberg, E., Berg, O. G. & Andersson, D. I. Experimental adaptation of Salmonella typhimurium to mice. Genetics 168, 1119–1130, doi: 10.1534/genetics.104.030304 (2004).
Giraud, A. et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291, 2606–2608 (2001).
Herr, A. J. et al. Mutator suppression and escape from replication error–induced extinction in yeast. PLoS Genet 7, e1002282, doi: 10.1371/journal.pgen.1002282 (2011).
Williams, L. N., Herr, A. J. & Preston, B. D. Emergence of DNA polymerase e antimutators that escape error-Induced extinction in yeast. Genetics 193, 751–770, doi: 10.1534/genetics.112.146910 (2013).
Fijalkowska, I. J. & Schaaper, R. M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc. Natl. Acad. Sci. USA 93, 2856–2861 (1996).
Zeyl, C., Mizesko, M. & de Visser, J. A. G. M. Mutational meltdown in laboratory yeast populations. Evolution 55, 909–917 (2001).
Daee, D. L., Mertz, T. M. & Shcherbakova, P. V. A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc. Natl. Acad. Sci. USA 107, 157–162, doi: 10.1073/pnas.0907526106 (2010).
Fijalkowska, I. J. & Schaaper, R. M. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J. Bacteriol. 177, 5979–5986 (1995).
Schaaper, R. M. & Cornacchio, R. An Escherichia coli dnaE mutation with suppressor activity toward mutator mutD5. J. Bacteriol. 174, 1974–1982 (1992).
Tröbner, W. & Piechocki, R. Selection against hypermutability in Escherichia coli during long term evolution. Mol. Gen. Genet. 198, 177–178 (1984).
Herr, A. J., Williams, L. N. & Preston, B. D. Antimutator variants of DNA polymerases. Crit. Rev. Biochem. Mol. Biol. 46, 548–570, doi: 10.3109/10409238.2011.620941 (2011).
Venkatesan, R. N., Hsu, J. J., Lawrence, N. A., Preston, B. D. & Loeb, L. A. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J. Biol. Chem. 281, 4486–4494 (2006).
Swan, M. K., Johnson, R. E., Prakash, L., Prakash, S. & Aggarwal, A. K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol 16, 979–986, doi: 10.1038/nsmb.1663 (2009).
Reha-Krantz, L. J. & Nonay, R. L. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J. Biol. Chem. 269, 5635–5643 (1994).
Li, L., Murphy, K. M., Kanevets, U. & Reha-Krantz, L. J. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae . Genetics 170, 569–580 (2005).
Schmitt, M. W., Matsumoto, Y. & Loeb, L. A. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie 91, 1163–1172, doi: 10.1016/j.biochi.2009.06.007 (2009).
Foster, P. L. Methods for determining spontaneous mutation rates. Methods Enzymol. 409, 195–213 (2006).
Zheng, Q. Methods for comparing mutation rates using fluctuation assay data. Mutat. Res. 777, 20–22, doi: 10.1016/j.mrfmmm.2015.04.002 (2015).
Zhou, Z. & Elledge, S. J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75, 1119–1127 (1993).
Zhao, X., Chabes, A., Domkin, V., Thelander, L. & Rothstein, R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20, 3544–3553 (2001).
Williams, L. N. et al. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc. Natl. Acad. Sci. USA, doi: 10.1073/pnas.1422948112 (2015).
Datta, A. et al. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol. Cell 6, 593–603 (2000).
Mertz, T. M., Sharma, S., Chabes, A. & Shcherbakova, P. V. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. USA, doi: 10.1073/pnas.1422934112 (2015).
Zhao, X., Muller, E. G. & Rothstein, R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340 (1998).
Chabes, A., Domkin, V. & Thelander, L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274, 36679–36683 (1999).
Fasullo, M., Tsaponina, O., Sun, M. & Chabes, A. Elevated dNTP levels suppress hyper-recombination in Saccharomyces cerevisiae S-phase checkpoint mutants. Nucleic Acids Res. 38, 1195–1203, doi: 10.1093/nar/gkp1064 (2010).
Lynch, M. Evolution of the mutation rate. Trends Genet. 26, 345–352, doi: 10.1016/j.tig.2010.05.003 (2010).
Kuchta, R. D., Benkovic, P. & Benkovic, S. J. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry (Mosc). 27, 6716–6725 (1988).
Fortune, J. M. et al. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 280, 29980–29987 (2005).
Nick McElhinny, S. A., Stith, C. M., Burgers, P. M. J. & Kunkel, T. A. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. M609591200, doi: 10.1074/jbc.M609591200 (2006).
Tran, H. T. et al. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol. Cell. Biol. 15, 5607–5617 (1995).
Stocki, S. A., Nonay, R. L. & Reha-Krantz, L. J. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J. Mol. Biol. 254, 15–28 (1995).
Hogg, M., Aller, P., Konigsberg, W., Wallace, S. S. & Doublie, S. Structural and biochemical investigation of the role in proofreading of a b hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 282, 1432–1444 (2007).
Hadjimarcou, M. I., Kokoska, R. J., Petes, T. D. & Reha-Krantz, L. J. Identification of a mutant DNA polymerase δ in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics 158, 177–186 (2001).
Sherman, F. In Part B: Guide to Yeast Genetics and Molecular and Cell Biology Vol. 350 Methods in Enzymology (eds Christine Guthrie & Gerald R. Fink ) 3–41 (Academic Press, 2002).
Boeke, J. D., LaCroute, F. & Fink, G. R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345–346 (1984).
Tong, A. H. & Boone, C. Synthetic genetic array analysis in Saccharomyces cerevisiae . Methods Mol. Biol. 313, 171–192 (2006).
Giot, L., Simon, M., Dubois, C. & Faye, G. Suppressors of thermosensitive mutations in the DNA polymerase δ gene of Saccharomyces cerevisiae . Mol. Gen. Genet. 246, 212–222 (1995).
Rose, M. D., Novick, P., Thomas, J. H., Botstein, D. & Fink, G. R. A. Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60, 237–243 (1987).
Gietz, R. D. & Sugino, A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 (1988).
Stulemeijer, I. J. et al. Dot1 binding induces chromatin rearrangements by histone methylation-dependent and -independent mechanisms. Epigenetics Chromatin 4, 2, doi: 10.1186/1756-8935-4-2 (2011).
Lee, Y. D., Wang, J., Stubbe, J. & Elledge, S. J. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 32, 70–80 (2008).
Huang, M., Zhou, Z. & Elledge, S. J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595–605 (1998).
Acknowledgements
We acknowledge Tom Walsh for assistance with whole genome sequencing and Ranga Venkatesan for the kind gift of the YCplac111pol3-01,L612M plasmid. We thank Brad Preston for valuable discussions. We thank Mitchell Lee for careful reading of the manuscript. This project was supported by R03 AG037081 and a Nathan Shock Center Junior Faculty Award, NIH/NIA P30 AG013280-18, from the National Institute On Aging Fund. Additional funding for AJH was provided by a Hitchings-Elion Fellowship from the Burroughs Wellcome Fund and R01GM118854 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute On Aging, the National Institute of General Medical Sciences, the National Institutes of Health, or the Burroughs Wellcome Fund.
Author information
Authors and Affiliations
Contributions
D.G.D. designed the study, performed experiments, and analyzed data. J.M.F., K.E., I.D., and S.R.K. performed experiments and analyzed data. A.J.H. designed the study, performed experiments, analyzed data, and wrote the manuscript; all authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Dennis, D., McKay-Fleisch, J., Eitzen, K. et al. Normally lethal amino acid substitutions suppress an ultramutator DNA Polymerase δ variant. Sci Rep 7, 46535 (2017). https://doi.org/10.1038/srep46535
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep46535
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.