Abstract
Melanoma of the uveal tract accounts for approximately 5% of all melanomas and represents the most common primary intraocular malignancy. Despite improvements in diagnosis and more effective local therapies for primary cancer, the rate of metastatic death has not changed in the past forty years. In the present study, we made use of bioinformatics to analyze the data obtained from three public available microarray datasets on uveal melanoma in an attempt to identify novel putative chemotherapeutic options for the liver metastatic disease. We have first carried out a meta-analysis of publicly available whole-genome datasets, that included data from 132 patients, comparing metastatic vs. non metastatic uveal melanomas, in order to identify the most relevant genes characterizing the spreading of tumor to the liver. Subsequently, the L1000CDS2 web-based utility was used to predict small molecules and drugs targeting the metastatic uveal melanoma gene signature. The most promising drugs were found to be Cinnarizine, an anti-histaminic drug used for motion sickness, Digitoxigenin, a precursor of cardiac glycosides, and Clofazimine, a fat-soluble iminophenazine used in leprosy. In vitro and in vivo validation studies will be needed to confirm the efficacy of these molecules for the prevention and treatment of metastatic uveal melanoma.
Similar content being viewed by others
Introduction
Uveal melanoma is the most common primary intraocular cancer, and after the skin, the uveal tract is the second most common location for melanoma1. However, cutaneous and uveal melanomas are different in terms of biology, natural history and response to chemotherapies2.
The 5-year survival rate is 50–70% and about 50% of patients develop metastases within a median of 36 months, almost exclusively to the liver, with a median survival of 6 months after metastases3.
The most frequent chromosomal abnormalities in uveal melanoma are loss of chromosome 3 and gains of 8q and 6p. Patients with chromosome 3 loss undergo the worst prognosis, whereas those with chromosome 6 (6p) gain have the best outcomes. Mutations in G-protein-α subunits GNAQ or GNA11 are observed in ≥80% of primary uveal melanomas and inactivating BAP1 mutations are found in approximately 50% of all cases, most frequently in metastatic disease. Mutations that are associated with a less aggressive behavior are those in splicing factor 3B subunit 1 (SF3B1) and eukaryotic translation initiation factor 1A, X-linked (EIF1AX)4.
Local radiation therapy, using brachytherapy or alternatively, charged-particle and proton-beam radiation, is the most common approach4,5,6. Enucleation remains the only option for very large tumors.
Despite improved understanding of the disease, the overall survival (OS) rate has not increased since the 1970 s. Indeed, once uveal melanoma has metastasized to distant organs, the disease is resistant to current chemotherapies. Distant metastasis are infrequent at the time of initial presentation, occurring in <5% of patients. For patients who develop metastasis, there is yet no standard of care. Dacarbazine, has been used for uveal melanoma, but efficacy is limited. Other chemotherapeutics, i.e. temozolomide, cisplatin, treosulfan, fotemustine and various combinations have been tested in uveal melanoma with disappointing results. A few adjuvant studies have been performed in uveal melanoma to prevent metastatic disease. However, no significant effects on metastasis free survival or OS benefit have been obtained, up to date4.
In the present study, we made use of bioinformatics to analyze the data obtained from three public available microarray datasets on uveal melanoma in attempt to identify novel putative chemotherapeutic options for the metastatic disease.
Material and Methods
Microarray dataset selection and meta-analysis
The NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/)7 was used to identify suitable microarray datasets comparing localized vs. metastatic uveal melanomas. Three datasets were included in the study GSE221388- GSE736529, and GSE44295, that included whole-genome transcriptomic data from primary melanoma cells obtained upon eye enucleation (Table 1). The following information were extracted from each of the studies that were selected: GEO accession; sample type; platform; numbers of patients and controls; and gene expression data. Briefly, the GSE22138 dataset included 28 samples of non-metastatic tumor and 35 samples from enucleated patients having distant metastasis. Patients were included in the group of localized tumors if no metastasis were observed in 36 months of follow-up. The GSE73652 dataset included 5 samples of non-metastatic tumor and 8 samples from enucleated patients having distant metastasis. Metastasis-free cases were included only if there was >6 years follow-up. No detailed clinical data are available for GSE44292, that included 32 samples from patients with localized disease and 24 sample from patients with metastasis. Illumina platforms were pre-processed using Bead-array prior to quantile normalization (Dunning et al., 2007), while Affymetrix platforms were pre-processed and quantile normalized using the robust multiarray average (RMA). The datasets were uploaded to INMEX (http://www.inmex.ca/INMEX)10, and the data annotated by converting probe ID to Entrez IDs. For each probe-set, intensity values were auto-scaled and a data integrity check was performed prior to the meta-analysis stage. Batch effects were corrected using the “ComBat” function. A random effects model of effect size (ES) measure was used to integrate gene expression patterns from the three datasets. The random effects model presumes that different studies present substantial diversity, and evaluates between-study variance as well as within-study sampling error. Genes with FDR < 0.01 were identified as Differentially Expressed (DE) and selected for further analysis.
Gene Ontology (GO) and Drug Prediction Analysis
Functional relationships among DE gene were obtained from GeneMania (http://genemania.org/ http://genemania.org/)11. GeneMANIA searches publicly available genomics and proteomics data, including data from gene and protein expression profiling studies and primary and curated molecular interaction networks and pathways, to find related genes. The network weighting method is ‘Gene-Ontology (GO) based weighting, Biological Process based’. This weighting method assumes the input gene list is related in terms of biological processes (as defined by GO).
The PANTHER (protein annotation through evolutionary relationship) classification system12 (http://www.pantherdb.org/) was used to classify input genes according to their function (PANTHER protein class). PANTHER is a comprehensive system that combines gene function, ontology, pathways and statistical tools that enable to analyze large-scale, genome-wide data from sequencing, proteomics or gene expression experiments.
The L1000CDS2 was used to identify potential chemotherapeutics for metastatic uveal melanoma. L1000CDS2 enables to find L1000 small molecule signatures that match input gene signatures. The L1000 mRNA gene-expression dataset is generated as part of the Library of Integrated Network-based Cellular Signatures (LINCS) project, a NIH Common Fund program. LINCS aims to profile the molecular effects of small molecules on human cells13. When gene lists are submitted to L1000CDS2, the search engine compares the input gene lists to the DE genes computed from the LINCS L1000 data and returns the top 50 matched signatures. The result score is the overlap between the input DE genes and the signature DE genes divided by the effective input. The effective input is the length of the intersection between the input genes and the L1000 genes. L1000CDS2 currently covers chemically perturbed gene expression profiles from 62 cell-lines and 3,924 small molecules. Also, L1000CDS2 allows to predict effective drug combinations by comparing every possible pair among the top 50 signatures and computing the potential synergy for each pair.
Results
Meta-analysis of gene expression in metastatic uveal melanoma
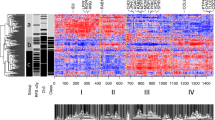
Three GEO data sets were identified for the following analysis (Table 1). These datasets consisted of primary uveal melanoma data, and included a total of 67 metastatic tumors and 65 localized primary tumors. Details of the individual studies are presented in Table 1. Figure 1A shows a Principal Component Analysis of the three separate microarray datasets included in the meta-analysis.
A total of 64 genes were identified, which were consistently modulated in metastatic tumors. Among these 64 DE genes, 29 were upregulated and 35 were downregulated. A list of the significant upregulated and downregulated genes is shown in Tables 2 and 3, respectively. An heatmap showing the 64 DE genes is presented in Fig. 1B. The functional relationships among the up- and down-regulated genes, respectively, are presented in Fig. 2A,B. Among the upregulated genes, the top three represented protein classes were: Transferase, which included phosphatases (CDC25B and IMPA1) and proteases (IDE and TYSND1); Enzyme Modulator, including G protein modulators (SGSM2, SRGAP2 and SIPA1L2) and the protease inhibitor, HPN; Nucleic Acid Binding, i.e. the DNA helicase, CHD7, and the DNA ligase, LIG1 (Fig. 2C).
(A) Functional network of the significant upregulated genes obtained from the meta-analysis of GSE73652, GSE44295 and GSE22138; (B) Functional network of the significant downregulated genes obtained from the meta-analysis of GSE73652, GSE44295 and GSE22138; (C) Protein class analysis for the significant upregulated genes obtained from the meta-analysis of GSE73652, GSE44295 and GSE22138; (D) Protein class analysis for the significant downregulated genes obtained from the meta-analysis of GSE73652, GSE44295 and GSE22138.
Among the downregulated genes, the top three protein classes represented were: Nucleic Acid Binding, that included DNA binding proteins (H2AFY2, NDN and MBNL1) and RNA binding proteins (ANG, EIF4A2 and EIF4A3); Hydrolase, including the proteases, ABHD6 and LTA4H, and the lipase, PLCD1; Enzyme Modulator, including the G protein modulator, PLCD1, the kinase modulator, HOOK1, and the protease inhibitor, VWA5A (Fig. 2D).
Prediction of novel chemotherapeutics for metastatic uveal melanoma
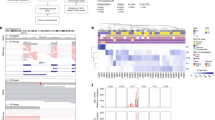
The L1000CDS2 web-based utility was used to predict small molecules and drugs targeting the metastatic uveal melanoma gene signature. Figure 3 shows a clustergram with the top ranked L1000 perturbations (e.g. those with most anti-similar signatures). The complete list of predicted chemotherapeutics is presented as Table 4. Several of these drugs are FDA-approved and already used in the clinic or tested in clinical trials, as indicated in Table 4. Among the predicted chemotherapeutics, the most represented classes were: histone deacetylase inhibitors (that included HDAC6 inhibitor ISOX, BRD-K13810148, Trichostatin A and Vorinostat), and anti-infectious/parasitic drugs (i.e., Clofazimine, Erythromycin ethylsuccinate, Demeclocycline and Quinacrine hydrochloride). The top identified drugs were the following: BRD-K07220430 (Cinnarizine), an anti-histaminic drug used for motion sickness, was predicted for its ability to downregulate CHAC1 and to upregulate MBNL1, LPAR6, PLSCR4, NDN, ABHD6, ZSCAN18 and ZBTB20; Digitoxigenin, for its ability to downregulate CDC25B, IDE, INTS8 and MTDH, while upregulating F11R, ID2, and RAB11FIP1; clofazimine, a fat-soluble iminophenazine used in leprosy, which is able to downregulate CDC25B, CHAC1, and SHC1, and to upregulate ABHD6, PLOD2, PLSCR4, ZBTB20, ZSCAN18. Accordingly, the top three most promising drug combination found were: BRD-K07220430 and Digitoxigenin; Digitoxigenin and OSSK_645683; BRD-K07220430 and HDAC6 inhibitor ISOX (for the complete list, see Table 5).
Discussion
Melanoma of the uveal tract accounts for 5% of all melanomas and, with an incidence of about 6 cases per million person–years, represents the most common primary intraocular malignancy14,15. Despite improvements in diagnosis and more effective local therapies for primary cancer, the rate of metastatic death has not changed in the past forty years15.
Several genes and pathways have been identified to be involved in the progression and metastasis of uveal melanoma, such as c-Met16, Hepatocyte Growth Factor (HGF)16,17, Insulin-like Growth Factor-1 Receptor (IGF-1R)18, the CXCL12-CXCR4 pathway17,19, VEGF20, Mda-9/syntenin21 and the PTP4A3 phosphatase8. However, despite increasing knowledge in the biology of uveal melanoma, no therapeutic strategies has been found to be effective. Metastatic uveal melanoma is resistant to current systemic chemotherapy and, to date, no clear role has been established for chemotherapy in several clinical trials, that reported objective response rates <20%4. Single-agent chemotherapies (e.g., dacarbazina, fotemustine, paclitaxel, temozolomide, camptothecin, treosulfan) or combination chemotherapies (e.g. gemcitabine/treosulfan, cisplatin/gemcitabine/treosulfan, carboplatin/paclitaxel/sorafenib) have shown poor response rates and immunotherapy with ipilimumab, antiangiogenetic treatment strategies using bevacizumab combined with interferon-α2b, temozolomide, or aflibercept have not proven to be superior to chemotherapy22.
In order to prevent metastatic disease, several adjuvant studies are also being conducted using ipilimumab, sunitinib, valproic acid, and crizotinib for high-risk patients4. Adjuvant treatment with bacillus Calmette-Guerin and interferon-α, as well as, intra-arterial hepatic infusion of fotemustine, have been previously studied in an effort to reduce the occurrence of liver metastasis, but none of these studies has demonstrated significant improvement in metastasis free survival or OS4.
The limited efficacy of current chemotherapies proves the medical need for more effective treatment strategies in metastatic uveal melanoma. In the present study, a meta-analysis of three whole-genome microarray datasets on primary tumor samples from the enucleated eyes of patients with localized or metastatic disease, has been performed in order to investigate the genes associated to the metastatic properties of uveal melanoma and to identify potential chemotherapeutic strategies to be used in metastatic disease.
It was recently developed a 15-gene qPCR-based assay that discriminate between primary uveal melanomas that have a low metastatic risk and a high metastatic risk 23. This platform is currently used in a College of American Pathologists (CAP)-accredited Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory on fine needle aspiration samples and on formalin-fixed specimens23. A few of the genes screened in this assay correspond to those identified by the current analysis, namely ID2 and LTA4H, among the downregulated genes. Some of the other DE genes, share similar function to those included in the assay, i.e. EIF4A2 and EIF4E2 (to EIF1B).
As regards the putative drugs here identified, some of them are already in clinical use, such as BRD-K07220430 (Cinnarizine)24, clofazimine25, mesoridazine besylate26, erythromycin ethylsuccinate27; other are in preclinical development (e.g. HDAC6 inhibitor ISOX)28 or do not have any known pharmacological target, e.g. OSSK_645683. Interestingly, some of the identified drugs are histone deacetylase inhibitors (such as, HDAC6 inhibitor ISOX, BRD-K13810148, Trichostatin A and Vorinostat), that have already been shown efficacy on uveal melanoma preclinical models29,30,31,32,33,34,35,36. Klisovic and collaborators have shown that the histone deacetylase inhibitor, Depsipeptide (FR901228), inhibits proliferation and induces apoptosis in primary and metastatic human uveal melanoma cell lines35, as well as it is able to inhibit in vitro uveal melanoma cell lines migration via downregulation of Matrix MetalloProteinases 2, 9 and Membrane Type-1/MMP (MMP-2, MMP-9 and MT-1/MMP) and the upregulation of Tissue Inhibitors of Matrix MetalloProteinases 1 and 2 (TIMP-1 and TIMP-2)34. Chen and collaborators have shown that microRNA-137 and microRNA-124a act as a tumor suppressors in uveal melanoma and could be successfully silenced by using the histone deacetylase inhibitor, trichostatin A32,36. Landreville and collegues have shown that in three uveal melanoma cell lines (92.1, OCM1A, and Mel202), Trichostatin A was able to reduce the fraction of viable cells and increase the proportion of cells undergoing apoptosis33. Also, it was shown that Quisinostat (a Class I and II histone deacetylase inhibitor) inhibited the migration and proliferation of 92.1 and OMM2.3 cell lines in zebrafish xenograft embryos30. In addition, the Class III-specific HDAC inhibitor, Tenovin-6, was shown to be able to eradicate cancer stem cells in 92.1 and Mel 270 cells29. Venza in 201431, reported that the Class-I histone deacetylase inhibitor, MS-275, due to its ability to reduce c-FLIP expression, is able to increase TRAIL-induced cell death in uveal melanoma cell lines. MS-275, is currently tested in a Phase 1 study for the treatment of patients with Refractory Solid Tumors, including intraocular melanoma, or Lymphomas (NCT00020579). Vorinostat, a small molecule inhibitor of class I and II histone deacetylases, is currently tested in two Phase 2 Study on Ocular Melanoma With Extraocular Extension, Recurrent Uveal Melanoma and Grade IV Metastatic Uveal Melanoma (NCT00121225, NCT 01587352).
The utility of the present data may reside primarily in the potential identification of effective adjuvant treatments. In light of the fact that there is no therapy for metastatic disease, the most promising strategy to improve survival is to treat patients in an adjuvant setting. Although there might be non- metastasizing melanomas, which never metastasize, even if not treated, however the intra-tumoral genetic heterogeneity suggests an ongoing evolutionary tumoral process37. Adjuvant interventions could settle up the uncertainty about the effect of ocular treatment on survival, since some patients might unnecessarily sacrifice their visus in the hope of a longer life-expectancy; while other patients with small melanomas might succumb due to metastasis because treatment has been delayed. Presently, there is no adjuvant approach that improves outcome, but the impact of ocular treatment, in terms of therapeutic benefit, has ethical implications.
The present study has several advantages but also limitations. It represents, to date, the largest meta-analysis comparing metastatic vs. non metastatic uveal melanomas, encompassing whole-genomic data from a total of 132 patients. A strict and rigorous statistics has been applied to data in order to sort out the most relevant genes characterizing the spreading of tumor to the liver. Among the upregulated genes, the highest effect size was detected for JPH1, encoding for Junctophilin-1, a component of junctional complexes between the plasma membrane and endoplasmic/sarcoplasmic reticulum, providing functional cross-talk between the cell surface and intracellular calcium release channels (http://www.genecards.org/cgi-bin/carddisp.pl?gene=JPH1). Among the significant downregulated genes, the gene with the highest effect size was PLSCR4, encoding for the Phospholipid Scramblase 4, which has been found to be strongly expressed in the neuropil of malignant gliomas and in cytoplasm of liver cancers, colorectal cancers and malignant melanomas (http://www.proteinatlas.org/ENSG00000114698-PLSCR4/cancer). Comparing to the original analysis, the PTP4A3 phosphatases and the cancer-testis antigen, PRAME, were dropped out because of too little statistical significance. In addition, although the role of the PKC, MAPK and PI3K/AKT/mTOR signaling cascades has been extensively examined in uveal melanoma38,39,40, however, whole genome transcriptomic analysis may fail to detect their modulation, because of the primary contribution related to post-transcriptional modifications. Also, our analysis was very statistical stringent (FDR < 0.01), and this may account for the differences from previously published work1. Furthermore, the discrepancies with previous analysis may be due to the enlargement of the number of samples which could influence DE genes detection as respect to smaller sample groups.
Investigating the genes found to be significantly modulated in the present analysis may help to identify the molecular mechanisms involved in the liver metastasis of uveal melanoma, and may allow the identification of novel pharmacological targets for the prevention and treatment of liver involvement. Moreover, our study was aimed at predicting putative pharmacological schemes to apply as adjuvant or curative therapy, making use of the Library of Integrated Network-based Cellular Signatures (LINCS) project database. The repurposing of drugs currently approved for use in the clinical setting may expedite the design of phase II-III clinical trials, being cost-effective and reducing the risks associated with early stages of drug development41. The highest ranking scores were found for Cinnarizine, Digitoxigenin and Clofazimine. Cinnarizine has already been shown to be effective in vitro against lymphoma and multiple myeloma42 and to inhibit melanogenesis in mouse B16 melanoma cells43, and Clofazimine was found effective in preclinical models of pancreatic ductal adenocarcinoma and triple-negative breast cancer44,45.
However, this study has also limitations. Despite the practicality of this bioinformatic approach, there are drawbacks to point out. First, the efficacy of a drug is more complex than the simple match of expression profiles. Drugs have to reach tissue-specific concentrations to exert an effect, and the route and timing of administration is essential for the drug to be effective and to limit side effects. A preliminary in vitro testing on both primary and established cancer cell lines will be required before running pilot phase II clinical trials. On the other hand, for successful result of the clinical trials, the appropriate selection and recruitment of patients will be also key to evaluate the potential activity of the compounds in the clinical setting.
Additional Information
How to cite this article: Fagone, P. et al. Identification of novel chemotherapeutic strategies for metastatic uveal melanoma. Sci. Rep. 7, 44564; doi: 10.1038/srep44564 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Zhang, Y., Yang, Y., Chen, L. & Zhang, J. Expression analysis of genes and pathways associated with liver metastases of the uveal melanoma. BMC Med. Genet. 15, 29 (2014).
McLaughlin, C. C. et al. Incidence of noncutaneous melanomas in the U.S. Cancer 103, 1000–7 (2005).
Shields, J. A., Shields, C. L., De Potter, P. & Singh, A. D. Diagnosis and treatment of uveal melanoma. Semin. Oncol. 23, 763–7 (1996).
Chattopadhyay, C. et al. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer 122, 2299–2312 (2016).
Diener-West, M. et al. The COMS randomized trial of iodine 125 brachytherapy for choroidal melanoma, III: initial mortality findings. COMS Report No. 18. Arch. Ophthalmol. (Chicago, Ill. 1960) 119, 969–82 (2001).
Egger, E. et al. Eye retention after proton beam radiotherapy for uveal melanoma. Int. J. Radiat. Oncol. Biol. Phys. 55, 867–80 (2003).
Clough, E. & Barrett, T. In 93–110, doi: 10.1007/978-1-4939-3578-9_5 (2016).
Laurent, C. et al. High PTP4A3 Phosphatase Expression Correlates with Metastatic Risk in Uveal Melanoma Patients. Cancer Res. 71, 666–674 (2011).
Field, M. G. et al. PRAME as an Independent Biomarker for Metastasis in Uveal Melanoma. Clin. Cancer Res. 22, 1234–1242 (2016).
Xia, J. et al. INMEX–a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res. 41, W63–70 (2013).
Zuberi, K. et al. GeneMANIA Prediction Server 2013 Update. Nucleic Acids Res. 41, W115–W122 (2013).
Mi, H., Poudel, S., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342 (2016).
Duan, Q. et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 42, W449–60 (2014).
Weis, E. et al. Management of uveal melanoma: a consensus-based provincial clinical practice guideline. Curr. Oncol. 23, e57–64 (2016).
Singh, A. D., Turell, M. E. & Topham, A. K. Uveal Melanoma: Trends in Incidence, Treatment, and Survival. Ophthalmology 118, 1881–1885 (2011).
Peruzzi, B. & Bottaro, D. P. Targeting the c-Met signaling pathway in cancer. Clin. Cancer Res. 12, 3657–60 (2006).
Di Cesare, S. et al. Expression and migratory analysis of 5 human uveal melanoma cell lines for CXCL12, CXCL8, CXCL1, and HGF. J. Carcinog. 6, 2 (2007).
All-Ericsson, C. et al. Insulin-like growth factor-1 receptor in uveal melanoma: a predictor for metastatic disease and a potential therapeutic target. Invest. Ophthalmol. Vis. Sci. 43, 1–8 (2002).
Scala, S. et al. CXC chemokine receptor 4 is expressed in uveal malignant melanoma and correlates with the epithelioid-mixed cell type. Cancer Immunol. Immunother. 56, 1589–1595 (2007).
Franco, R. et al. "CXCR4-CXCL12 and VEGF correlate to uveal melanoma progression". Front . Biosci. (Elite Ed). 2, 13–21 (2010).
Gangemi, R. et al. Mda-9/Syntenin Is Expressed in Uveal Melanoma and Correlates with Metastatic Progression. PLoS One 7, e29989 (2012).
Buder, K., Gesierich, A., Gelbrich, G. & Goebeler, M. Systemic treatment of metastatic uveal melanoma: review of literature and future perspectives. Cancer Med. 2, 674–86 (2013).
Harbour, J. W. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol. Biol. 1102, 427–40 (2014).
Scholtz, A.-W., Ilgner, J., Loader, B., Pritschow, B. W. & Weisshaar, G. Cinnarizine and dimenhydrinate in the treatment of vertigo in medical practice. Wien. Klin. Wochenschr. 128, 341–347 (2016).
Cholo, M. C., Mothiba, M. T., Fourie, B. & Anderson, R. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J. Antimicrob. Chemother. dkw426, doi: 10.1093/jac/dkw426 (2016).
Aguilar, S. J. An open study of mesoridazine (Serentil) in chronic schizophrenics. Dis. Nerv. Syst. 36, 484–9 (1975).
Rogers, G. B., Bruce, K. D., Martin, M. L., Burr, L. D. & Serisier, D. J. The effect of long-term macrolide treatment on respiratory microbiota composition in non-cystic fibrosis bronchiectasis: an analysis from the randomised, double-blind, placebo-controlled BLESS trial. Lancet Respir. Med. 2, 988–996 (2014).
Butler, K. V. et al. Rational Design and Simple Chemistry Yield a Superior, Neuroprotective HDAC6 Inhibitor, Tubastatin A. J. Am. Chem. Soc. 132, 10842–10846 (2010).
Dai, W., Zhou, J., Jin, B. & Pan, J. Class III-specific HDAC inhibitor Tenovin-6 induces apoptosis, suppresses migration and eliminates cancer stem cells in uveal melanoma. Sci. Rep. 6, 22622 (2016).
Van der Ent, W. et al. Modeling of Human Uveal Melanoma in Zebrafish Xenograft Embryos. Investig. Opthalmology Vis. Sci. 55, 6612 (2014).
Venza, I., Visalli, M., Oteri, R., Teti, D. & Venza, M. Class I-specific histone deacetylase inhibitor MS-275 overrides TRAIL-resistance in melanoma cells by downregulating c-FLIP. Int. Immunopharmacol. 21, 439–446 (2014).
Chen, X. et al. MicroRNA-124a Is Epigenetically Regulated and Acts as a Tumor Suppressor by Controlling Multiple Targets in Uveal Melanoma. Investig. Opthalmology Vis. Sci. 54, 2248 (2013).
Landreville, S. et al. Histone Deacetylase Inhibitors Induce Growth Arrest and Differentiation in Uveal Melanoma. Clin. Cancer Res. 18, 408–416 (2012).
Klisovic, D. D. et al. Depsipeptide inhibits migration of primary and metastatic uveal melanoma cell lines in vitro: a potential strategy for uveal melanoma. Melanoma Res. 15, 147–53 (2005).
Klisovic, D. D. et al. Depsipeptide (FR901228) inhibits proliferation and induces apoptosis in primary and metastatic human uveal melanoma cell lines. Invest. Ophthalmol. Vis. Sci. 44, 2390–8 (2003).
Chen, X. et al. Epigenetics, MicroRNAs, and Carcinogenesis: Functional Role of MicroRNA-137 in Uveal Melanoma. Investig. Opthalmology Vis. Sci. 52, 1193 (2011).
Dopierala, J., Damato, B. E., Lake, S. L., Taktak, A. F. G. & Coupland, S. E. Genetic Heterogeneity in Uveal Melanoma Assessed by Multiplex Ligation-Dependent Probe Amplification. Investig. Opthalmology Vis. Sci. 51, 4898 (2010).
Nabil, A.-A. et al. Upcoming translational challenges for uveal melanoma. Br. J. Cancer 113, 1249–1253 (2015).
Sagoo, M. S., Harbour, J. W., Stebbing, J. & Bowcock, A. M. Combined PKC and MEK inhibition for treating metastatic uveal melanoma. Oncogene 33, 4722–4723 (2014).
Gaudi, S. & Messina, J. L. Molecular Bases of Cutaneous and Uveal Melanomas. Patholog. Res. Int. 2011, 1–8 (2011).
Chong, C. R. & Sullivan, D. J. New uses for old drugs. Nature 448, 645–646 (2007).
Schmeel, L. C. et al. In vitro efficacy of cinnarizine against lymphoma and multiple myeloma. Anticancer Res. 35, 835–41 (2015).
Chang, T.-S. & Lin, V. C.-H. Melanogenesis Inhibitory Activity of Two Generic Drugs: Cinnarizine and Trazodone in Mouse B16 Melanoma Cells. Int. J. Mol. Sci. 12, 8787–8796 (2011).
Zaccagnino, A. et al. Tumor-reducing effect of the clinically used drug clofazimine in a SCID mouse model of pancreatic ductal adenocarcinoma. Oncotarget, doi: 10.18632/oncotarget.11299 (2014).
Koval, A. V. et al. Anti-leprosy drug clofazimine inhibits growth of triple-negative breast cancer cells via inhibition of canonical Wnt signaling. Biochem. Pharmacol. 87, 571–578 (2014).
Author information
Authors and Affiliations
Contributions
The research idea was proposed by A.R., A.L. and M.R. The experiments were designed and performed by P.F. and F.N. The data were analyzed and results interpreted by R.C., G.L., D.A., S.B., R.D.P. and M.L. P.F. and F.N. wrote the main manuscript. The revision and editing of the manuscript was performed by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Fagone, P., Caltabiano, R., Russo, A. et al. Identification of novel chemotherapeutic strategies for metastatic uveal melanoma. Sci Rep 7, 44564 (2017). https://doi.org/10.1038/srep44564
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44564
This article is cited by
-
Genome-wide CRISPR knockout screening identified G protein pathway suppressor 2 as a novel tumor suppressor for uveal melanoma metastasis
Journal of Cancer Research and Clinical Oncology (2023)
-
miR-204 suppresses uveal melanoma cell migration and invasion through negative regulation of RAB22A
Functional & Integrative Genomics (2023)
-
Establishment and validation of five autophagy-related signatures for predicting survival and immune microenvironment in glioma
Genes & Genomics (2022)
-
Identification and validation of signal recognition particle 14 as a prognostic biomarker predicting overall survival in patients with acute myeloid leukemia
BMC Medical Genomics (2021)
-
Prospective validation in epithelial tumors of a gene expression predictor of liver metastasis derived from uveal melanoma
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.