Abstract
DNA replication is tightly regulated to constrain the genetic material within strict spatiotemporal boundaries and copy numbers. Bacterial plasmids are autonomously replicating DNA molecules of much clinical, environmental and biotechnological interest. A mechanism used by plasmids to prevent over-replication is ‘handcuffing’, i.e. inactivating the replication origins in two DNA molecules by holding them together through a bridge built by a plasmid-encoded initiator protein (Rep). Besides being involved in handcuffing, the WH1 domain in the RepA protein assembles as amyloid fibres upon binding to DNA in vitro. The amyloid state in proteins is linked to specific human diseases, but determines selectable and epigenetically transmissible phenotypes in microorganisms. Here we have explored the connection between handcuffing and amyloidogenesis of full-length RepA. Using a monoclonal antibody specific for an amyloidogenic conformation of RepA-WH1, we have found that the handcuffed RepA assemblies, either reconstructed in vitro or in plasmids clustering at the bacterial nucleoid, are amyloidogenic. The replication-inhibitory RepA handcuff assembly is, to our knowledge, the first protein amyloid directly dealing with DNA. Built on an amyloid scaffold, bacterial plasmid handcuffs can bring a novel molecular solution to the universal problem of keeping control on DNA replication initiation.
Similar content being viewed by others
Introduction
The molecular mechanisms of DNA replication in Gram-negative bacteria have been the subject of intense research for four decades. In the case of most plasmids, a plasmid-encoded protein (Rep) triggers replication in a regulated way1. In RepA from the Pseudomonas pPS10 plasmid replicon2, its N-terminal winged-helix dimerization domain (WH1) is structurally remodelled upon binding to DNA, resulting in the transformation of stable transcriptional repressor dimers into metastable replication-competent monomers3. In the plasmid replication origin (oriV) RepA monomers assemble the initiation complex at specific directly repeated sequences (iterons)4,5. Once replicated, two iteron-containing plasmid DNA molecules get coupled in a ‘handcuffed’ complex through interactions mediated by origin-bound Rep molecules, sterically hindering premature replication rounds6,7,8,9,10. In previous work, we characterized that the handcuffed complexes responsible for negative regulation of pPS10 replication were mediated by RepA monomers coupled through their WH1 domains, albeit involving a distinct interface to that found in the repressor RepA dimers11.
Tracking the molecular basis of the intrinsic tendency of RepA towards aggregation, we found that monomers of the isolated RepA-WH1 domain assemble into amyloid fibres in vitro12,13. A plasmid-specific dsDNA sequence acts as allosteric effector of amyloidosis12,14. When fused to a fluorescent protein marker, RepA-WH1 behaves as a proteinopathic, vertically transmissible (from mother cell to daughter cells) prionoid in Escherichia coli thus enabling bacteria as a model system for approaching protein amyloidosis15,16. We have recently described a monoclonal antibody (B3h7) specific for an oligomeric conformation of RepA-WH1 on pathway towards building amyloid fibres17. B3h7 thus overcame limitations imposed by the poor reactivity of RepA-WH1 towards commercially available anti-amyloid antibodies (such as A11 and OC)17. Using B3h7, we discovered that pre-amyloidogenic RepA-WH1 oligomers assemble at the bacterial nucleoid17, as expected from the DNA-promoted amyloidogenesis of the protein in vitro12,14.
Protein amyloids, in their fibrillar or oligomeric aggregated states, are infamous as the causative agents of human degenerative proteinopathies spanning from Alzheimer’s, Parkinson’s, Huntington’s, prion diseases and amyotrophic lateral sclerosis to dialysis-related amyloidosis and type-II diabetes18,19. However, work performed on yeast prions20 and bacterial biofilms21 have clearly shown that amyloids can also be functional, i.e. provide microorganisms with quickly selectable epigenetic, gain of function phenotypes22. Here we explore the link between regulation of pPS10 replication by RepA-mediated origin handcuffing and DNA-promoted RepA-WH1 amyloidosis. We have found that the regulatory RepA handcuffs actually are, to our notice, the first intracellular functional amyloids found in bacteria or involved in DNA replication.
Results
Amyloidogenic RepA mediates handcuffed plasmid complexes in vitro
An important open question is whether the basis of two phenomena involving the WH1 domain, i.e. handcuffing of RepA-iteron complexes and amyloidosis of RepA-WH1, were or not a single process (amyloidogenesis), albeit in naturally functional or synthetic proteinopathic contexts, respectively. With such purpose, we have used B3h7, a monoclonal antibody specific for on-pathway amyloidogenic RepA-WH1 oligomers17, as a probe to explore a possible contribution of amyloidogenesis to handcuffing.
We reconstructed in vitro the complexes between full-length RepA and plasmid DNA molecules carrying the pPS10 operator or iteron sequences11 and then performed Western/dot-blotting (Fig. 1) or immuno-electron microscopy (iEM) (Fig. 2) using the B3h7 antibody. α-WH1, a polyclonal antibody specific for RepA-WH1 but not its conformation17, was also tested in these assays as a control. We thus surveyed if RepA adopts an amyloid structure in two distinct functional assemblies: i) RepA dimers bound at the repA operator inverted repeat; and ii) RepA monomers titrated on the oriV iterons, either as handcuffed complexes or as the intermediates of binding.
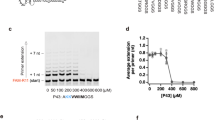
Antibodies used recognize RepA-WH1 either irrespective of its conformation (polyclonal α-WH1) or as amyloidogenic oligomers (monoclonal B3h7)17. Complexes assembled in vitro between full-length RepA and ≈1 kb plasmid restriction fragments carrying either the repA operator (pUC-oriV0) (a) or the four pPS10 iterons (pUC-oriV4) (b), were resolved in agarose gels (left) and then Western-blotted to membranes before incubation with the indicated antibodies (right). Duplicates of the same samples were serially diluted and then directly dot-blotted to membranes before incubation with the same antibodies (bottom). Plasmid schemes highlight the distinct RepA conformations involved in transcriptional repression (black circles), DNA replication initiation (chequered ovals) and handcuffing (grey ovals)3,4,5,11. B3h7 preferentially recognizes the high molecular weight RepA-oriV complexes at the conditions in which two origin fragments appeared handcuffed in trans11 (Fig. 2, right), pointing to an amyloidogenic nature for such regulatory complex.
Image galleries of in vitro reconstituted complexes between full length RepA and plasmids carrying the pPS10 repA operator (left) or the four iterons array at oriV (right). Linearized plasmid molecules were incubated with RepA (a–c) and antibodies (b,c) before negative staining and Pt-shadowing. (a) Besides free plasmid molecules, oligomeric RepA-DNA complexes are evident (white arrows) for both types of sequences. (b) iEM with the primary polyclonal α-WH1 antibody and a secondary gold-conjugated antibody (Au-IgG, 10 nm Ø; black arrows). RepA is found at the core of the assemblies identified in (a), either as bound to the operator (left) or as origin-bound or handcuffed (right). (c) iEM with the monoclonal anti-oligomeric amyloid antibody B3h7 and secondary Au-IgG (black arrows). B3h7 does not efficiently recognize the operator-bound RepA dimers (white arrows; the single black arrow marks a background antibody molecule) (left), whereas B3h7 identifies (black arrows) the oligomers assembled by the RepA monomers at the handcuffed complexes as amyloidogenic (right).
The titration of RepA on operator (Fig. 1a) and oriV iteron (Fig. 1b) DNA sequences, present in plasmids that had been sliced into pieces through multiple restriction digestion, showed specific mobility shifts (native EMSA) only for the fragment carrying the relevant pPS10 sequences. In the case of RepA binding to the iterons, Western blotting with the B3h7 antibody revealed an intense hybridization signal solely for the highest molecular weight complex, located close to the well of the gel, but not for any of the intermediate monomers binding cooperatively4 to the four iterons at oriV (Fig. 1b). On the contrary, the signal generated at the well for the samples including the operator was significantly less intense than that observed for the iterons, i.e. some protein aggregation happened but no signal showed up for the specific complex between RepA dimers and DNA (Fig. 1a). The control α-WH1 antibody recognized every complex in which a RepA molecule was taking part, either as a dimer at repA operator (Fig. 1a) or as the individual monomers binding to each iteron (Fig. 1b). Dot-blot analysis of serial dilutions of the titration points for both types of DNA sequences revealed that samples including RepA-iteron complexes (Fig. 1b) were labelled with B3h7 up to higher dilutions than those with RepA-operator complexes (Fig. 1a) and, importantly, only at the titration points in which handcuffing complexes were evident in EMSA, whereas α-WH1 recognized both kinds of samples to a similar extent. The differences observed between the hybridization patterns for both antibodies speak to their distinct specificities, as recently reported17: either for an oligomeric and amyloidogenic conformation of RepA (B3h7) or for multiple peptide epitopes distributed across RepA regardless the conformation or association state of the protein (α-WH1). In summary, these approaches indicate that the largest complexes built by RepA at the oriV iterons, corresponding to handcuffed origin molecules11, involve amyloidogenic oligomers, whereas the individual RepA monomer-iteron and RepA dimer-operator complexes do not.
Complementary iEM analysis of the individual RepA-DNA complexes, as reconstituted on linearized plasmid molecules (Fig. 2a), indicated that while the α-WH1 polyclonal antibody recognized any RepA particle bound to DNA, either RepA dimers at the operator (Fig. 2b, left) or RepA monomers at the iterons (Fig. 2b, right), labelling by the amyloidogenic oligomer-specific monoclonal B3h7 antibody (Fig. 2c) showed a clear preference for the latter (Fig. 2c, right). A quantitative analysis of the iEM images revealed that B3h7 labelled all the handcuffed complexes, but it also recognized half of the single uncoupled RepA-oriV particles (Fig. 3). Whether the latter were from dissociation of handcuffed complexes during handling of the samples cannot be excluded. In contrast, and compatible with the findings in the Western and dot-blot assays (Fig. 1), B3h7 barely labelled one-fourth of the RepA-operator complexes (Fig. 3). By focusing at a nanometer scale on individual complexes, iEM with the B3h7 probe provides unambiguous evidence for the existence of an amyloidogenic structure for the bridge made of RepA that holds handcuffed plasmids together (see scheme in Fig. 1b).
Particles were classified according to the assembly type of the DNA molecules (single/linear vs. double/handcuffed) and to the distinct reactivity of RepA towards the α-WH1 and B3h7 antibodies (see Fig. 2). RepA complexes with the iteron tandem repeats, with a preference for the handcuffed state, were identified as amyloidogenic by the B3h7 probe. Data were compiled from three independent reconstitutions plus EM-imaging.
Amyloidogenic RepA handcuffing at the nucleoid of P. aeruginosa cells carrying pPS10
The B3h7 antibody combined with the nanometer-range resolution of electron microscopy were successful in detecting foci of the amyloidogenic RepA-WH1 prionoid at the nucleoid of E. coli cells17. Therefore, the same approach was also the choice for imaging the whole RepA protein, not just its N-terminal WH1 domain, as expressed from its natural promoter in a pPS10 replicon undergoing controlled RepA-dependent DNA replication in P. aeruginosa cells, the natural host for the plasmid (Fig. 4). As for the aggregated amyloid precursors of the RepA-WH1 prionoid in E. coli cells17, in P. aeruginosa B3h7 labelled the nucleoid territory (Fig. 4b), whereas signal from the polyclonal α-WH1 antibody, besides the nucleoid, extended also to the cytoplasm probably pointing to RepA molecules unbound to DNA (Fig. 4a). As expected from their high specificity for RepA, none of the two antibodies targeted bacteria not carrying a pPS10 replicon (Fig. 4c). Labelling by B3h7 of both the nucleoid of bacteria carrying pPS10 (Fig. 4b) and the handcuffs reconstituted in vitro (Figs 1b and 2c, right) provide strong support to the proposal that the regulatory nucleoprotein complexes involved in the control of plasmid DNA replication are built on an amyloidogenic RepA backbone.
(a) iEM performed with the α-WH1 polyclonal antibody on ultra-thin sections (≈90 nm) of P. aeruginosa PAO1024 cells bearing a functional pPS10 plasmid replicon. RepA molecules (arrows pointing to Au particles) appeared sparsely distributed across the whole bacterial cells. (b) iEM with the anti-amyloid B3h7 antibody probe on cells from the same bacterial cultures shown in (a). Amyloidogenic RepA oligomers (arrows) were located as clusters inside the nucleoid (N, green sectors), resembling the amyloid precursors of the RepA-WH1 prionoid previously identified at the E. coli nucleoid17. (c) Negative control: plasmid-free bacterial cells probed with the α-WH1 and B3h7 antibodies; arrows, background Au particles.
Discussion
The findings reported here integrate amyloids in the complex conformational transactions experienced by RepA protein in a key process for the biology of plasmid extrachromosomal mobile genetic elements in Gram-negative bacteria: i) DNA (operator)-bound dimers repress repA gene transcription; ii) DNA (iteron)-bound monomers initiate DNA replication; and iii) by means of handcuffing, amyloidogenic oligomers inhibit post-replicative origin firing. A previous study11 established that handcuffing complexes involve RepA monomers coupled as pseudo-dimers through a surface in WH1 that includes the C-terminus of helix α2, which is distinct from the dimerization interface found in soluble and operator-bound RepA dimers3. Since the key amyloidogenic stretch in RepA-WH1 is also found at the C-terminus of α212, the findings reported here are fully compatible with a functional amyloid being involved in the control of pPS10 replication (Fig. 5). An amyloid protein core confers a high stability to the handcuffed complexes, providing a basis for the known requirement of the DnaK-DnaJ-GrpE chaperone triad in disassembling Rep handcuffs to allow for further plasmid replication rounds8,10. Interestingly, DnaK modulates the amyloidogenesis of the RepA-WH1 prionoid in vivo by shifting the equilibrium between an acutely and a mildly cytotoxic transmissible conformations (strains) of the protein towards the latter16.
(a) RepA in negative control of DNA replication (handcuffing)11. (b) RepA-WH1 in the polymerization of prionoid fibres13. Starting with soluble and stable dimers, dsDNA-promoted amyloidogenesis would drive the WH1 domain, in both scenarios and through aggregation-prone metastable protein monomers, into two distinct amyloid structures: WH1 domains paired in trans through a head-to-head interface (β1-β1), resulting in inhibition of DNA replication (a)11, or head-to-tail interactions involving a second, yet undefined interface (β1–β2), which build fibres with indefinite length (b)17. The factor limiting the span of RepA complexes in handcuffing (a) is the specific binding of the protein to the four iteron DNA direct repeats, as determined by the C-terminal WH2 domain3,4,5 that both stabilizes the nucleoprotein complex and shields the second amyloidogenic interface (β2, in white) avoiding subsequent polymerization.
The understanding of plasmid partition23, i.e. the molecular mechanism assuring the equal distribution of plasmid copies between daughter cells upon division, has recently advanced in a substantial way24,25. Partition of low copy-number plasmids relies on centromere-like sequences (parS), a parS-binding protein (ParB) and an ATP hydrolysing protein (ParA) which couples the partition complex to the nucleoid, then spread the plasmids to distal locations through diffusion ratchet26 or DNA looping27 mechanisms. In all cases studied so far, interactions involving ParA and ParB are mediated through α-helical interfaces24,25, not by β-sheets, as proposed here for RepA handcuffing. It is likely that handcuffing of plasmids is the basis for the amyloidogenic patches found in the nucleoid of bacteria carrying pPS10 (Fig. 3b). Plasmids lacking its own partition module but carrying sequences that match the centromere-like parS locus still become associated to the bacterial nucleoid for stable segregation by the ParAB proteins encoded at the chromosome28. This is likely the case for pPS10, because its replicon includes sequences (e.g.: C521CTTCCATGGGGAAGG536)2 bearing similarity to the consensus parS in P. aeruginosa29. Beyond its role in replication control, whether amyloid-mediated handcuffing has any effect on plasmid partition deserves further exploration, since the stable Rep-iteron complexes have been recently found to constrain ParB spreading30. Although speculative, it would be sufficient that at least one of the plasmids bridged through handcuffing would stay attached through its parS sites to the chromosome to achieve efficient partition of a plasmid cluster.
CPEB protein leads the list of functional amyloids dealing with mRNA, contributing to establish memory in neurons30,31. Analogously, the handcuffed RepA oligomer might be the foundational member of a class of amyloids modulating the dynamics of DNA and genomes.
Methods
Electrophoretic mobility-shift assays (EMSA)
Plasmids pUC-oriV0 (repA operator) and pUC-oriV4 (pPS10 four iterons)11 were digested with NdeI, AclI and AlwNI, to generate four fragments of which the largest one (≈ 1 kb) included the relevant sequence probes (see Fig. 1), and then purified through the FastPlasmid Mini Kit (5Prime, Fisher). Concentrations of the DNA probes were determined after gel electrophoresis, using calibrated MW standards and the Quantity-One software (Bio-Rad). Each binding reaction included 37 ng of the relevant dsDNA probe, acting the other three fragments in the incubation mixture as controls for non-specific binding. Purified histidine-tagged RepA-WT11 was pre-treated with 1 M urea at 4 °C for 15 min, to assure a monodisperse dimeric state for the protein, and then titrated on the dsDNA probes in a final volume of 10 μL (binding buffer: 0.05 M NaCl, 0.025 M Hepes pH 8, 5% glycerol, 0.1 mg mL−1 BSA). After 15 min on ice, samples were loaded in 1.5% agarose-0.25xTBE gels and electrophoresis performed at 100 V for 2 h at 4 °C. Gels were stained with Gel-Red (Biotium) and fluorescence emission images captured using a Gel-Doc 2000 (Bio-Rad).
Western and dot-blot assays
α-WH1 and B3h7 antibodies were obtained and purified as previously described17. Western blot assays were performed after EMSA by transferring the agarose gels to PVDF membranes in a Trans-Blot cell (Bio-Rad) at 16 V (400 mA), 4 °C for 15 h in 1 × TAE buffer supplemented with 0.025% SDS. For dot-blots, nitrocellulose membranes (0.45 μm ø pore; Bio-Rad) were set in a Bio-Dot microfiltration device (Bio-Rad). Wells were pre-equilibrated twice by flowing 200 μL of binding buffer (no BSA added). 72 ng of RepA in complex with DNA, replicas of the samples assembled for EMSA (see above), were serially diluted (2-fold steps) in the same buffer (200 μl final volume) and immediately spotted under gravity flow. In both assays, blotted membranes were then blocked, for 1 h at room temperature, with 5% skimmed powder milk in Tris-buffered saline buffer (pH 7.0) containing 0.01% Tween-20 (TBS-T) and probed for 2 h at room temperature with the primary antibodies (B3h7 1:3,000; or α-WH1 1:1,000) in TBS-T. The membranes were then washed three times with TBS-T and incubated for 1 h with the appropriate (mouse or rabbit, respectively) HRP-conjugated secondary antibodies (1:10,000). After three additional washes with TBS-T, chemiluminiscent detection was performed on X-ray films with the ECL Prime kit (GE Healthcare).
Electron microscopy
TEM/iEM of in vitro assembled RepA-DNA complexes: 40 ng of histidine-tagged RepA-WT were added to 40 ng of NdeI-linearized pUC-oriV0 or pUC-oriV4 (see above)11 in 10 μl of 200 mM KCl, 7 mM Mes (pH 6.0), 1.5 mM DTT, 3% glycerol, 1 mM ATP and incubated for 10 min at 4 °C. Glutaraldehyde was then supplied to 0.2% and cross-linking reaction proceeded for 10 min before quenching, by adding 2 μl of 0.5 M glycyl-glycine (pH 7.4) for 10 extra min. The reaction mixes were incubated with the primary antibodies (α-WH1 or B3h7, 0.6 ng.μl−1) for 30 min at room temperature and then diluted to 40 μl with TBT (100 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2). Samples were gel-filtrated through a TBT-Sephacryl S-500 column (GE Healthcare) and peak fractions were pooled and clarified by centrifugation (800× g, for 5 min). The Au-conjugated secondary antibodies (either anti-mouse or anti-rabbit, 1:40) were incubated with the samples for 30 min at room temperature. Reactions were further processed to clean unbound antibodies through the gel-filtration column as above, but equilibrated in 10 mM triethanolamine-HCl pH 7.5, 10 mM MgCl2. Samples were sequentially adsorbed on mica, stained with 2% uranyl acetate, platinum shadowed and carbon coated, before examination of the specimens in a FEI CM100 electron microscope, as described11.
iEM of bacterial cells: Cultures of P. aeruginosa PAO1024 carrying pRG14, a pPS10 replicon derivative encoding repA and oriV 2, were harvested in late exponential phase (OD600 = 1.0). Cells were fixed, embedded in resin, sliced, sequentially incubated on the grids with the primary antibodies (0.2 ng μl−1) and the secondary Au-conjugated antibodies and stained with uranyl acetate, as described in15,16,17. Specimens were examined in a JEOL JEM-1230 electron microscope, as indicated15,16,17.
Additional Information
How to cite this article: Molina-García, L. et al. Functional amyloids as inhibitors of plasmid DNA replication. Sci. Rep. 6, 25425; doi: 10.1038/srep25425 (2016).
References
Giraldo, R. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: Combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 26, 533–554 (2003).
Nieto, C., Giraldo, R., Fernández-Tresguerres, E. & Díaz, R. Genetic and functional analysis of the replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae patovar savastanoi . J. Mol. Biol. 223, 415–426 (1992).
Giraldo, R., Fernández-Tornero, C., Evans, P. R., Díaz-Orejas, R. & Romero, A. A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat. Struct. Biol. 10, 565–571 (2003).
Díaz-López, T. et al. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 278, 18606–18616 (2003).
Díaz-López, T., Dávila-Fajardo, C., Blaesing, F., Lillo, M. P. & Giraldo, R. Early events in the binding of the pPS10 replication protein RepA to single iteron and operator DNA sequences. J. Mol. Biol. 364, 909–920 (2006).
Blasina, A., Kittell, B. L., Toukdarian, A. E. & Helinski, D. R. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl. Acad. Sci. USA 93, 3559–3564 (1996).
Chattoraj, D. K. Control of plasmid DNA replication by iterons: No longer paradoxical. Mol. Microbiol. 37, 467–476 (2000).
Park, K., Han, E., Paulsson, J. & Chattoraj, D. K. Origin pairing (‘handcuffing’) as a mode of negative control of P1 plasmid copy number. EMBO J. 20, 7323–7332 (2001).
Kunnimalaiyaan, S., Inman, R. B., Rakowski, S. A. & Filutowicz, M. Role of π dimers in coupling (“handcuffing”) of plasmid R6K’s γ ori iterons. J. Bacteriol. 187, 3779–3785 (2005).
Zzaman, S. & Bastia, D. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol. Cell 20, 833–843 (2005).
Gasset-Rosa, F. et al. Negative regulation of pPS10 plasmid replication: Origin pairing by zipping-up DNA-bound RepA monomers. Mol. Microbiol. 68, 560–572 (2008).
Giraldo, R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl. Acad. Sci. USA 104, 17388–17393 (2007).
Torreira, E. et al. Amyloidogenesis of bacterial prionoid RepA-WH1 recapitulates dimer to monomer transitions of RepA in DNA replication initiation. Structure 23, 183–189 (2015).
Gasset-Rosa, F., Maté, M. J., Dávila-Fajardo, C., Bravo, J. & Giraldo, R. Binding of sulphonated indigo derivatives to RepA-WH1 inhibits DNA-induced protein amyloidogenesis. Nucleic Acids Res. 36, 2249–2256 (2008).
Fernández-Tresguerres, M. E., Moreno-Díaz de la Espina, S., Gasset-Rosa, F. & Giraldo, R. A DNA-promoted amyloid proteinopathy in Escherichia coli . Mol. Microbiol. 77, 1456–1469 (2010).
Gasset-Rosa, F. et al. Direct assessment in bacteria of prionoid propagation and phenotype selection by Hsp70 chaperone. Mol. Microbiol. 91, 1070–1087 (2014).
Moreno-del Álamo, M., Moreno-Díaz de la Espina, S., Fernández-Tresguerres, M. E. & Giraldo, R. Pre-amyloid oligomers of the proteotoxic RepA-WH1 prionoid assemble at the bacterial nucleoid. Sci. Rep. 5, 14669 (2015).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Eisenberg, D. & Jucker, M. The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012).
Wickner, R. B. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae . Science 264, 566–569 (1994).
Chapman, M. R. et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002).
Si, K. Prions : What are they good for? Annu. Rev. Cell. Dev. Biol. 31, 149–169 (2015).
Vecchiarelli, A. G., Mizuuchi, K. & Funnell, B. E. Surfing biological surfaces: exploiting the nucleoid for partition and transport in bacteria. Mol. Microbiol. 86, 513–523 (2012).
Schumacher, M. A. et al. Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science 349, 1120–1124 (2015).
Volante, A. & Alonso, J. C. Molecular anatomy of ParA-ParA and ParA-ParB interactions during plasmid partitioning. J. Biol. Chem. 290, 18782–18795 (2015).
Hwang, L. C. et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 32, 1238–1249 (2013).
Graham, T. G. et al. ParB spreading requires DNA bridging. Genes Dev. 28, 1228–1238 (2014).
Kolatka, K., Witosinska, M., Pierechod, M. & Konieczny, I. Bacterial partitioning proteins affect the subcellular location of broad-host-range plasmid RK2. Microbiology 154, 2847–2856 (2008).
Bartosik, A. A., Lasocki, K., Mierzejewska, J., Thomas, C. M. & Jagura-Burdzy, G. ParB of Pseudomonas aeruginosa: Interactions with its partner ParA and its target parS and specific effects on bacterial growth. J. Bacteriol. 186, 6983–6998 (2004).
Sanchez, A. et al. Stochastic self-assembly of ParB proteins builds the bacterial DNA segregation apparatus. Cell Syst. 1, 163–173 (2015).
Si, K. et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia . Cell 115, 893–904 (2003).
Acknowledgements
We are indebted to Mercedes Carnota for her technical assistance with iEM. This work has been financed with grants from Spanish MINECO (CSD2009-00088 and BIO2012-30852).
Author information
Authors and Affiliations
Contributions
L.M.-G. performed the biochemical characterization of RepA amyloids. F.G.-R. and R.L. carried out the iEM on the reconstituted handcuffing complexes. M.M.-dA. prepared the B3h7 antibody and supervised its assay. M.E.F.-T. and S.M.-D.d.l.E. did the iEM studies with bacteria. R.G. conceived the project, designed the experiments with contributions from the other authors, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Molina-García, L., Gasset-Rosa, F., Moreno-del Álamo, M. et al. Functional amyloids as inhibitors of plasmid DNA replication. Sci Rep 6, 25425 (2016). https://doi.org/10.1038/srep25425
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep25425
This article is cited by
-
Emerging roles of the cellular prion protein (PrPC) and 37/67 kDa laminin receptor (RPSA) interaction in cancer biology
Cellular and Molecular Life Sciences (2023)
-
Revised role for Hfq bacterial regulator on DNA topology
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.