Abstract
Faithful DNA replication is a prerequisite for cell proliferation. Several cytological studies have shown that chromosome structures alter in the S-phase of the cell cycle. However, the molecular mechanisms behind the alteration of chromosome structures associated with DNA replication have not been elucidated. Here, we investigated chromatin structures and acetylation of specific histone residues during DNA replication using the meiotic nucleus of the fission yeast Schizosaccharomyces pombe. The S. pombe meiotic nucleus provides a unique opportunity for measuring the levels of compaction of chromatin along the chromosome in a defined orientation. By direct measurement of chromatin compaction in living cells, we demonstrated that decompaction of chromatin occurs during meiotic DNA replication. This chromatin decompaction was suppressed by depletion of histone acetyltransferase Mst1 or by arginine substitution of specific lysine residues (K8 and K12) of histone H4. These results suggest that acetylation of histone H4 residues K8 and K12 plays a critical role in loosening chromatin structures during DNA replication.
Similar content being viewed by others
Introduction
The eukaryotic genome is organized into chromatin. Faithful meiotic DNA replication is a prerequisite for transmitting the genome from parent to offspring. Several cytological studies using a variety of experimental methods have supported a model in which chromosome structures alter in the S-phase of the cell cycle1,2,3,4. Studies using a premature chromosome condensation technique provided the first visual evidence for decondensation of chromatin fibers in S-phase1,2. Analysis of replication fork dynamics in living cells also suggests that the sites of DNA synthesis are sites of decondensed chromatin4. Posttranslational modification of histones, such as acetylation of histones H3 and H4, has been reported to regulate DNA replication5,6,7 probably by affecting chromatin structures8. However, the molecular mechanisms underlying the DNA replication-associated alteration of chromatin structures remain unknown.
The meiotic nucleus in the fission yeast Schizosaccharomyces pombe provides a useful experimental system in which to study chromatin structures in a defined orientation of chromosomes (Fig. 1A). In this organism, the nucleus elongates and moves back and forth between the cell ends upon entering meiosis (Supplementary Fig. S1A) and telomeres remain clustered at the leading edge of the moving nucleus; this elongated nucleus is generally called a “horsetail” nucleus due to its shape9,10. This unique arrangement of chromosomes in the horsetail nucleus has allowed direct measurements of chromatin compaction in living cells11. In our previous study, we measured chromatin compaction to demonstrate the function of meiotic cohesin Rec8 in chromosome compaction during meiotic prophase11.
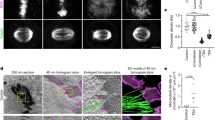
Measurements of chromatin compaction levels in living cells.
(A) Schematic diagram of the meiotic nucleus in S. pombe. In meiotic prophase, the telomeres of all three chromosomes are clustered at the spindle-pole body9,10. Under the pulling force of the horsetail movement, the meiotic nucleus exhibits an elongated morphology. (B) Schematic diagram of the lacO inserts (ade8 and ade1 loci) on chromosome II. (C) Live-cell imaging of chromosomal loci in the horsetail nucleus: wild type (WT), csn1Δ, csn1Δspd1Δ, ddb1Δ, rec8Δ and csn1Δrec8Δ. Telomeres were marked with Taz1-GFP; the ade8 locus was visualized using lacO/LacI-GFP. Scale bar indicates 5 μm. (D,F,H) A percentile rank plot of distances between the telomere and ade8 locus (D) or between the ade8 and ade1 loci (F, H) in the horsetail nucleus. (E,G,I) Statistical analysis of the distances measured in (D), (F) and (H). Means and standard deviations (SD) of the distances are shown. P values were calculated by Student’s t-test. A chromatin compaction ratio is defined as the length of DNA divided by the longitudinal length of chromatin. The 400-kb telomere-ade8 chromatin region contains 136 μm of DNA; the 1,000-kb ade8-ade1 chromatin region contains 340 μm of DNA
Taking an advantage of this characteristic nuclear morphology in S. pombe meiosis, we previously isolated a csn1 mutant in a screening of sporulation-deficient mutants that exhibits aberrant nuclear morphology12. The csn1+ gene encodes the subunit 1 of COP9 signalosome complex13, which is required for activity of the Pcu4-Ddb1Cdt2 E3 ubiquitin ligase complex14,15; COP9 signalosome complex and Pcu4-Ddb1Cdt2 ubiquitin ligase are required for degradation of Spd1, an inhibitor of ribonucleotide reductase (RNR). Because RNR activity is necessary to convert ribonucleotides to deoxyribonucleotides (dNTPs), deficiency of Csn1 causes stalled DNA replication due to inefficiency of dNTP synthesis14,15. In the current study, we measured chromatin compaction in the csn1 mutant and identified an association between histone H4 modification and chromatin compaction during DNA replication.
Results
Decompaction of chromatin occurs in mutants with stalled DNA replication
A csn1-deletion strain (csn1∆) exhibited a characteristic phenotype during nuclear movements12 similar to that reported in rec8∆ strain11: the tip of the horsetail nucleus moved back and forth within the cell while the bulk of the nucleus did not follow the movements (Supplementary Fig. S1B), implying that the chromatin was stretched under the pulling forces of horsetail movement. To quantify the degree of chromatin compaction, we measured the distances between two loci along the chromosome (the telomere, the ade8 locus or the ade1 locus) in the horsetail nucleus in living cells, using a telomere protein, Taz1, tagged with green fluorescent protein (Taz1-GFP) and the ade8 or ade1 locus tagged with the lacO/LacI-GFP system on chromosome II (Fig. 1B). The average distance between the telomere and ade8 locus in csn1∆ cells was 1.6-fold greater than in wild-type cells (Fig. 1C–E). The average distance between the ade8 locus and the ade1 locus in csn1∆ cells was also 1.5-fold greater than in wild-type cells (Fig. 1F,G), suggesting that chromatin is less compact in csn1∆ than in wild-type cells. The degree of chromatin compaction can be estimated by calculating a chromatin compaction ratio, which is defined as the length of DNA contained between two loci divided by the measured longitudinal length of the respective region of chromatin.
Because Csn1 is required for activity of the Pcu4-Ddb1Cdt2 ubiquitin ligase complex14,15, cells deleted for the ddb1+ gene (ddb1∆) are expected to exhibit phenotypes similar to csn1∆ cells. Thus, we examined nuclear morphology and measured the telomere-ade8 distance in ddb1∆ cells. As expected, the same phenotype of chromatin decompaction in the telomere-ade8 region was observed in ddb1∆ cells (Fig. 1C–E). Next we examined whether disruption of the spd1+ gene (spd1∆) could suppress formation of the abnormal horsetail nucleus, because the spd1∆ mutation is known to suppress meiotic arrest in csn1∆ and ddb1∆ cells15. The Spd1 protein is an inhibitor of RNR and degradation of Spd1 through ubiquitin pathway involving Csn1 and Ddb1 is required for the effective synthesis of dNTPs upon reaching the S phase. Results showed that deletion of the spd1+ gene suppressed chromatin decompaction in csn1∆ cells, as indicated by measurements of the telomere-ade8 distance (Fig. 1C–E). Taken together, these results suggest that decompaction of chromatin observed in the csn1∆ or ddb1∆ mutant is due to stalled DNA replication caused by insufficiency of dNTPs.
Since it is known that the loss of Rec8 causes chromatin decompaction in meiotic prophase11, we also measured the telomere-ade8 distance in rec8∆ and csn1∆ rec8∆ cells (Fig. 1C–E). The telomere-ade8 distance in rec8∆ was similar to that in csn1∆ and ddb1∆ cells (Fig. 1D,E). On the other hand, the distance in the csn1∆ rec8∆ double mutant was greater than in each single mutant, indicating that the chromosomes were more extended in rec8∆ under stalled DNA replication. These results suggest that stalled DNA replication has an additive effect, independent of cohesin function, on chromatin decompaction.
Chromatin decompaction is suppressed by depletion of histone acetyltransferase Mst1
We considered that the observed chromatin decompaction might involve specific chromatin modifications. Since some histone acetylations are coupled with DNA replication16,17,18, we searched for a histone acetyltransferase (HAT) that affects chromatin decompaction in the horsetail nucleus in csn1∆ cells. For this purpose, we constructed strains in which each of five non-essential HAT genes (hat1+, elp3+, gcn5+, rtt109+ and mst2+) was disrupted, or an essential HAT, Mst1, was conditionally depleted in the csn1∆ background and examined whether stretched morphology of the horsetail nucleus was suppressed in these strains. Whereas disruption of the non-essential HAT genes did not affect stretching of the horsetail nucleus (Supplementary Fig. S2A), depletion of Mst1 suppressed it in csn1∆ cells. Mst1, tagged with GFP-fused degron protein IAA17-GFP, was depleted using an improved-AID (i-AID) system19 (see Methods). Depletion of Mst1-IAA17-GFP was confirmed by fluorescence microscopy. To quantify the chromatin compaction levels, we measured the ade8-ade1 distance in wild-type and csn1∆ cells under conditions of Mst1 depletion. The ade8-ade1 distance was distinctly shortened in csn1∆ cells as a result of Mst1 depletion, while it was slightly shortened in wild-type cells (Fig. 1H; summarized in Fig. 1I). This observation implies that histone acetylation catalyzed by Mst1 is responsible for chromatin decompaction in csn1∆ cells.
Acetylation of histone H4 is necessary for chromatin decompaction during DNA replication
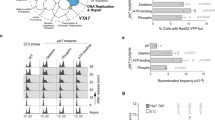
As it is known that Mst1 catalyzes histone H4 acetylation in S. pombe20, we examined the acetylation of lysine residues (K5, K8, K12 and K16) of histone H4 during meiosis. Acetylation of histone H3-K56, which was reported as a replication-coupled histone modification17, was used as a positive control. The K5, K8, K12 and K16 residues of histone H4 were acetylated for 100–180 min after the induction of meiosis, which corresponds to the period of DNA replication (Fig. 2).
Acetylation of histone H4 during meiotic S phase.
Haploid h– pat1-114 cells were cultured in nitrogen-free medium (EMM2-N) overnight at 26 °C. After 20 h, the culture was shifted to 34 °C to induce meiosis. (A) Flow cytometry of DNA contents during meiosis. Cells were sampled at the indicated time points for analysis. (B) Histone acetylation during meiosis. The cell extracts were prepared at the indicated time points and analyzed by immunoblotting. Actin was used as a loading control. Asynchronous cells (as) were obtained from preculture in YES medium.
To identify the amino acid residues of histone H4 responsible for chromatin decompaction, we substituted each of four candidate lysine residues to arginine (K5R, K8R, K12R and K16R). S. pombe has three copies of the histone H3-H4 gene cassette (hht1+-hhf1+, hht2+-hhf2+ and hht3+-hhf3+). To express a single copy of the mutated histone H4 gene, we mutated the hhf2 gene in the background of hht1-hhf1 hht3-hhf3 double deletion (see Methods). Expression of histone H4-K8R or -K12R partially suppressed chromatin decompaction in csn1∆ cells, whereas expression of histone H4-K5R or -K16R did not (Supplementary Fig. S1C). These results were confirmed by measurements of the ade8-ade1 distance: decompaction of chromatin in csn1∆ cells was suppressed by histone H4-K8R or -K12R, but not by histone H4-K5R or -K16R (Fig. 3A,B). These results indicate that the decompaction of chromatin requires acetylation of K8 and K12 residues of histone H4. Although Mst1 also catalyzes Pht1 (histone H2A.Z) acetylation in S. pombe21, deletion of the pht1+ gene did not suppress stretching of the horsetail nucleus in csn1∆ (Supplementary Fig. S2B), suggesting that histone H2A.Z is not involved in chromatin decompaction in these cells.
Chromatin decompaction and HU sensitivity in histone H4 mutants.
Percentile rank plot of the ade8-ade1 distance in csn1∆ cells with the indicated mutation of histone H4 (orange). The plot for csn1∆ expressing wild-type histone H4 (blue) was used as a control for all panels. (B) Statistical analysis of the distances measured in (A). (C) Percentile rank plot of the ade8-ade1 distance in meiotic S phase and in meiotic prophase in the wild-type background. (D,E) Percentile rank plot of the ade8-ade1 distance in meiotic S phase (D) and meiotic prophase (E) in cells expressing wild-type and mutant histone H4. (F) Statistical analysis of the distances measured in (C), (D) and (E). Means and standard deviations (SD) of the distances are shown. P values were calculated by Student’s t-test.
Since acetylation of histone H4 is involved in chromatin decompaction in csn1∆ cells, chromatin might be loosened during meiotic DNA replication when histone H4 is acetylated. To test this possibility, we measured the ade8-ade1 distance during S phase and prophase in living meiotic cells. Meiotic DNA replication occurs approximately at the beginning of the nuclear movements12,22,23. To distinguish S-phase cells from prophase cells, we used PCNA tagged with mCherry (mCherry-Pcn1) as a marker for DNA replication12,24. Cells containing the elongated horsetail nucleus with mCherry-Pcn1 foci represent S-phase cells and those without mCherry-Pcn1 foci represent meiotic prophase cells. In wild-type cells, the ade8-ade1 distance was longer in S-phase cells than in meiotic prophase cells (Fig. 3C), indicating that chromatin decompaction indeed occurs during meiotic DNA replication. Cells expressing histone H4-K5R or -K16R showed distance profiles similar to wild-type cells in both S phase and prophase (Fig. 3D,E; summarized in Fig. 3F). In contrast, cells expressing histone H4-K8R or -K12R exhibited significant shortening of the ade8-ade1 distance in S phase but not in prophase (Fig. 3D,E; summarized in Fig. 3F), suggesting that chromatin decompaction through histone H4 acetylation at K8 and K12 is specific to S phase. Furthermore, cells expressing histone H4 with mutations of both K8R and K12R (H4-K8R/K12R) exhibited more distinct shortening of the ade8-ade1 distance in S phase in comparison to the single mutations (Fig. 3D). Thus, we conclude that acetylation of histone H4 at K8 and K12 is critical for loosening of chromatin structures during DNA replication.
Acetylation of histone H4 at K8 and K12 residues is essential for vegetative growth
To consider the possibility that the expression of mutant histone H4 affects the duration of the meiotic S phase, we measured the duration of meiotic DNA replication using GFP-Pcn1 in the histone H4-K8R/K12R mutant. The results showed that meiotic DNA replication was completed within the same duration in these mutant cells in which chromatin decompaction was not observed during meiotic DNA replication (Fig. 4A). In addition, there is no significant difference in spore viability between wild type and the histone H4-K8R/K12R mutant (Fig. 4B). Thus, the decompaction of chromatin is not absolutely necessary for meiotic DNA replication.
Acetylation of histone H4 at K8 and K12 residues is essential for vegetative growth.
(A) Duration of meiotic S-phase in wild-type and histone H4-K8R/K12R cells. This experiment was performed as previous report12. Error bars represent the standard deviation. (B) Viability of vegetative cells and spores of wild-type and histone H4-K8R/K12R cells. Error bars represent the standard deviation. (C) HU sensitivity in cells expressing wild-type and mutant histone H4. Dilution series (1/5 dilution) of cell suspensions were spotted on complete medium (YES) with or without 6 mM HU and grown for three days at 30 °C.
In contrast, vegetative cells expressing histone H4-K8R/K12R exhibited slow growth on a YES plate. Thus, we measured vegetative viability of these mutant cells. The viability of these mutant cells was only 57.5% compared with 93.6% in wild-type cells (Fig. 4B). It also should be noted that this mutant showed supersensitivity to hydroxyurea (HU), an inhibitor of RNR to decrease dNTP synthesis (Fig. 4C). These results suggest that acetylation of those histone H4 residues may necessary for vegetative growth in order to circumvent problems caused by stalled DNA replication. The difference in viability between vegetative cells and spores is an intriguing question and remains to be elucidated.
Discussion
In this study, our direct measurements of chromatin compaction in the meiotic horsetail nucleus of living S. pombe cells demonstrated that chromatin decompaction occurs during meiotic DNA replication. Our results also show that this decompaction is due to DNA replication-coupled acetylation of histone H4 by Mst1. DNA replication-coupled histone acetylation has also been reported in vegetative cells16,17,18; in addition, Mst1 interacts with MCM proteins25, suggesting that Mst1-mediated histone acetylation occurs during DNA replication in the mitotic cell cycle as well as in meiosis. Histone H4 acetylation may affect S-phase progression through regulation of gene expression. In fact, bromodomain-containing proteins bind to acetylated histones26,27 and regulate gene expression of some genes28,29. However, our measurements in living cells directly demonstrate that histone H4 acetylation alters chromatin compaction levels without affecting S-phase progression.
We also identified the K8 and K12 residues of histone H4 as critical acetylation sites for chromatin decompaction during DNA replication (Fig. 5). In addition, the histone H4-K8R/K12R mutant is hypersensitive to HU, suggesting that acetylation of K8 and K12 of histone H4 play an important role in the response to DNA replication stress. However, a previous report showed that a histone H4 mutant bearing four substitutions (K5R, K8R, K12R and K16R) had no effect on HU sensitivity20. This apparent discrepancy is probably due to the presence of two copies of the wild-type histone H4 gene in the reported mutant strains whereas our H4 mutant strains bear a mutated histone H4 gene as the sole histone H4 gene.
Interestingly, overproduction of Clr6, which catalyzes the deacetylation of histone H4, prevents phosphorylation of replication checkpoint Cds1 under DNA replication stress30. Therefore, defects in recovery of stalled DNA replication may be the result of impaired Cds1 function in the H4-K8R/K12R mutant. However, it has also been reported that high acetylation levels of histone H4 has a negative role in stabilization of replication forks in the absence of Cds120. Thus, histone H4 acetylation plays multiple roles in the response to DNA replication stress.
Our results showed that the histone H4-K16R mutation did not affect chromatin decompaction during DNA replication. However, acetylation of H4-K16 is considered important for regulating the formation of higher-order chromatin structures31,32,33,34,35. The crystal structure of the nucleosome and in vitro chromatin assembly assays indicate residues 14–19 of histone H4 in the nucleosome interact with the acidic region on the surface of the adjacent nucleosome31,32,33. In addition, acetylation of histone H4-K16 prevents formation of 30-nm chromatin fibers34 while acetylation at histone H4-K5, K8 and K12 together only have a small additive effect on formation of the 30-nm fiber35. The difference between these in vitro assays and our in vivo assays may be because replication-coupled decompaction of chromatin occurs at a different order of chromatin organization instead of 30 nm fiber formation. In fact, the apparent compaction ratios of meiotic prophase chromatin are approximately 100, which is more compact than the ratio of 42 for the 30 nm fiber. Thus, the sites that are important for formation of 30 nm fibers may not be involved in the alteration of chromatin structures during DNA replication. Although further studies are required to address how acetylation of histone H4 regulates higher-order chromatin structures, our study clearly indicate a link between histone H4-K8/K12 acetylation and chromatin decompaction during DNA replication.
Methods
Cell culture and media
For routine culture, YES and EMM2 were used as complete medium and synthetic medium, respectively36. To induce meiosis, h90 mating-type cells were suspended in EMM2 without a nitrogen source (EMM2-N) and the suspension was spotted on ME agar plates. When h+ and h– mating-type cells were used to induce meiosis, both strains were mixed in EMM2-N and the suspension was spotted on ME.
Construction of strains and plasmids
The strains used in this study are listed in Table S1. Gene disruption and tagging with GFP, mRFP, or mCherry were performed as described12. The i-AID degron system was used for depletion of Mst1 protein19. To express skp1-AtTIR1-2NLS-9myc from the lys1 gene locus, a 4.4-kb NotI fragment containing a Padh15-skp1-AtTIR1-2NLS-9myc fragment was obtained from pMK10419 and a lys1-N marker fragment was PCR-amplified from the pCST3 plasmid23. Both fragments were cloned into pBluescript II SK (-) (Stratagene) and the resulting plasmid pBlue-lys1-AtTIR1 was used for transformation. The transformants were selected on EMM2 plates lacking lysine and used to tag Mst1 protein using IAA17-GFP. For IAA17-GFP tagging, the IAA17 gene was PCR-amplified from the HM1905 genome19 and integrated into the SalI site of pFA6-GFP (S65T)-kanMX637 using an In-Fusion HD Cloning kit (Takara). The resulting plasmid was used to tag the endogenous mst1 gene using a two-step PCR method38,39 and the constructed strain was used for Mst1-depletion experiments.
We constructed histone H4 mutant strains harboring only hht2-hhf2 in the background of hht1-hhf1 hht3-hhf3 double deletion as follows. To generate a mutated hht2-hhf2 gene fragment, the entire hht2-hhf2 locus (2.1 kb) was cloned by PCR from the wild-type genome (L968 strain). The fragment was inserted into a plasmid and a point mutation was introduced by site-directed mutagenesis using the PrimeSTAR Mutagenesis Basal Kit (Takara). After the mutation site was confirmed by sequencing, the resulting hht2-hhf2 mutant gene fragment was used for transformation. To construct histone mutant strains, strains from which hht2+-hhf2+ had been deleted with a ura4 marker in the background of hht1-hhf1 deletion (RK1021) or hht3-hhf3 deletion (HA1090-12D) were transformed with the mutant gene fragment. RK1021 and HA1090-12D were generated from YTP55440. Transformants were selected on YES agar containing 1 mg/mL 5-fluoroorotic acid (5-FOA). The mutations were confirmed by PCR and sequencing. Strains containing the mutated hht2-hhf2 allele were crossed to obtain progeny harboring the hht2-hhf2 mutant gene allele in the background of hht1-hhf1 deletion and hht3-hhf3 deletion.
Fluorescence microscopy
A DeltaVision fluorescence microscopy system (Applied Precision), which is based on an Olympus wide-field IX71 fluorescence microscope equipped with an oil-immersion objective lens (Plan Apo 60×; NA = 1.4; Olympus) and a CoolSNAP HQ2 CCD camera (Photometrics), in a temperature-control room, was used for imaging of the fission yeast cells41. Cells were grown on YES agar at 30 °C or 26 °C for the cdc10-129ts strain. To induce meiosis, the cells were transferred to an ME plate at 26 °C. After the formation of zygotes, the cells were suspended in EMM-N medium. Then, the cells were mounted on a 24 mm × 60 mm cover glass coated with 0.2% (w/v) lectin. Images were collected of 15 focal planes at 0.5 μm intervals. To measure the telomere-ade8 or ade8-ade1 distance, horsetail nuclei were observed in zygotes. The distance was measured only when the nucleus was moving straight in either direction (not making a turn). Time-lapse observation was performed as described12. Images were deconvolved with SoftWorx software (Applied Precision). The distance was measured using Priism software (University of California, San Francisco)42. Each trace was initiated at the center of the GFP focus.
Induction of meiosis in Mst1-depleted conditions
For Mst1-depletion experiments, a cdc10-129ts allele was used to arrest csn1∆ cells at G1 phase before meiosis induction43,44 because csn1∆ cells are difficult to arrest at the G1 phase by nitrogen starvation. cdc10-129ts csn1∆ cells were arrested for 4 h at the restrictive temperature of 36 °C to accumulate in G1. For the depletion of Mst1-IAA17-GFP protein, 0.5 mM auxin (1-naphthaleneacetic acid) (Nacalai Tesque) was added 2 h after the temperature shift to 36 °C. Then, the cells were mated on an auxin-containing EMM2-N agar plate at 26 °C. Depletion of Mst1-IAA17-GFP was confirmed by fluorescence microscopy.
Viability assay of vegetative cells and spores
Log-phase cells cultured in YES were used for assay of vegetative viability. For spore viability assay, log-phase cells were washed twice with EMM2-N. Then the cells were spotted on ME plate and incubate at 26 °C for 2 days. Asci were digested by glucuronidase (Sigma) at 26 °C overnight. Cells or spores were separated manually using a micromanipulator on a tetrad microscope. For each sample, 120 cells or spores were inoculated on YES plates and cultured to form colonies at 30 °C for 7 days. Viability was determined from the number of colonies formed out of the number of cells or spores inoculated. Means and standard deviations were obtained from three measurements.
Immunoblotting analysis
A total of 2.5 × 107 asynchronously growing cells or 5 × 107 G1-arrested cells were collected by centrifugation and the pellets were frozen at −80 °C. For immunoblotting, the cells were suspended in 800 μl ice-cold water containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and disrupted with 150 μl of 2 M NaOH. Then, the cells were fixed with 150 μl of 55% trichloroacetate. After centrifugation, the pellets were dissolved in 200 μl of sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 12.5 mM EDTA and 10% glycerol). Proteins in the extracts were separated by 15% SDS-PAGE and transferred to a PVDF membrane using a semidry system. The membranes were incubated for 1 h at room temperature in PBST buffer (10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, 1.76 mM KH2PO4 and 0.05% Tween-20) containing 5% skim milk. Then, the membranes were incubated with anti-actin antibody (Abcam), anti-H3K56 antibody (Millipore), anti-H4K5ac, anti-H4K8ac, anti-H4K12ac, anti-H4K16ac, or H4 antibody45 in PBST containing 1% skim milk at 4 °C overnight. After washing three times with PBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Millipore) in PBST containing 1% skim milk at room temperature for 1 h. The signals were visualized with ImmuneStar LD (Wako) and detected using Chemidoc MP imaging system (Bio-Rad).
Flow cytometry
A total of 1 × 107 cells were fixed with 70% ethanol and incubated with 50 μg/mL RNaseA in 500 μl of 50 mM sodium citrate (pH 7.5) for 4 h at 37°C and then stained with 0.5 μg/mL propidium iodide. DNA content of 1.5 × 105 cells was measured on a FACS Calibur instrument (BD Biosciences).
Additional Information
How to cite this article: Ruan, K. et al. Histone H4 acetylation required for chromatin decompaction during DNA replication. Sci. Rep. 5, 12720; doi: 10.1038/srep12720 (2015).
References
Johnson, R. T. & Rao, P. N. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 226, 717–722 (1970).
Hanks, S. K., Gollin, S. M., Rao, P. N., Wray, W. & Hittelman, W. N. Cell cycle-specific changes in the ultrastructural organization of prematurely condensed chromosomes. Chromosoma 88, 333–342 (1983).
Li, G., Sudlow, G. & Belmont, A. S. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140, 975–989 (1998).
Kuipers, M. A. et al. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol. 192, 29–41 (2011).
Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J. & Grunstein, M. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233 (2002).
Kaykov, A. & Nurse, P. The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 25, 391–401 (2015).
Luciano, P., Dehé, P. M., Audebert, S., Géli, V. & Corda, Y. Replisome function during replicative stress is modulated by histone h3 lysine 56 acetylation through Ctf4. Genetics 199, 1047–1063 (2015).
Bhaskara, S. et al. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin 6, 27 (2013).
Chikashige, Y. et al. Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273 (1994).
Ding, D.-Q., Chikashige, Y., Haraguchi, T. & Hiraoka, Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111, 701–712 (1998).
Ding, D.–Q. et al. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 174, 499–508 (2006).
Ruan, K. et al. Meiotic nuclear movements in fission yeast are regulated by the transcription factor Mei4 downstream of a Cds1-dependent replication checkpoint pathway. Genes Cells 20, 160–172 (2014).
Wei, N., Serino, G. & Deng, X.-W. The COP9 signalosome: more than a protease. Trends Biochem Sci 33, 592–600 (2008).
Liu, C. et al. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 17, 1130–1140 (2003).
Holmberg, C. et al. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19, 853–862 (2005).
Sobel, R. E., Cook, R. G., Perry, C. A., Annunziato, A. T. & Allis, C. D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92, 1237–1241 (1995).
Xhemalce, B. et al. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 282, 15040–15047 (2007).
Unnikrishnan, A., Gafken, P. R. & Tsukiyama, T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 17, 430–437 (2010).
Kanke, M. et al. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 12, 8 (2011).
Garabedian, M. V. et al. The Double-Bromodomain Proteins Bdf1 and Bdf2 Modulate Chromatin Structure to Regulate S-Phase Stress Response in Schizosaccharomyces pombe. Genetics 190, 487–500 (2012).
Kim, H.–S. et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 16, 1286–1293 (2009).
Bähler, J., Wyler, T., Loidl, J. & Kohli, J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121, 241–256 (1993).
Chikashige, Y., Kurokawa, R., Haraguchi, T. & Hiraoka, Y. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells 9, 671–684 (2004).
Meister, P., Taddei, A., Ponti, A., Baldacci, G. & Gasser, S. M. Replication foci dynamics: replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J. 26, 1315–1326 (2007).
Gomez, E. B., Nugent, R. L., Laria, S. & Forsburg, S. L. Schizosaccharomyces pombe Histone Acetyltransferase Mst1 (KAT5) Is an Essential Protein Required for Damage Response and Chromosome Segregation. Genetics 179, 757–771 (2008).
Zeng, L. & Zhou, M. M. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513, 124–128 (2002).
Sanchez, R. & Zhou, M. M. The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Devel. 12, 659–665 (2009).
Stejskal, S., Tesařová, L. & Koutná, I. Mysterious role of H3K56ac in embryonic stem cells. Folia Biol. (Praha) 60 (Suppl 1), 71–75 (2014).
Nagarajan, S., Benito, E., Fischer, A. & Johnsen, S. A. H4K12ac is regulated by estrogen receptor-alpha and is associated with BRD4 function and inducible transcription. Oncotarget 6, 7305–7317 (2015).
Kunoh, T., Habu, T. & Matsumoto, T. Involvement of fission yeast Clr6-HDAC in regulation of the checkpoint kinase Cds1. Nucleic Acids Res 36, 3311–3319 (2008).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T. J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003).
Iwasaki, W. et al. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio. 3, 363–369 (2013).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Allahverdi, A. et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 39, 1680–1691 (2011).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 194, 795–823 (1991).
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Wach, A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 (1996).
Hayashi, A. et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells 14, 217–225 (2009).
Takayama, Y. & Takahashi, K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 35, 3223–3237 (2007).
Haraguchi, T. et al. Multiple-color fluorescence imaging of chromosomes and microtubules in living cells. Cell Struct. Funct. 24, 291–298 (1999).
Chen, H., Hughes, D. D., Chan, T. A., Sedat, J. W. & Agard, D. A. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116, 56–60 (1996).
Nurse, P., Thuriaux, P. & Nasmyth, K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146, 167–178 (1976).
Petersen, J., Nielsen, O., Egel, R. & Hagan, I. M. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111, 867–876 (1998).
Eberhart, A. et al. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 21, 535–554 (2013).
Acknowledgements
We thank Atsushi Matsuda for image processing; Yuko Takayama and Shigeaki Saitoh for providing the histone gene disruption strains; Da-Qiao Ding for providing the lacO/LacI-GFP strains; Mai Kanke for providing the strains and plasmids for the degron system; and Chizuru Ohtsuki for technical assistance. This work was supported by JSPS KAKENHI Grants (26440098 to H.A., 21370094 to T.H. and 20114002, 26116511 to Y.H.).
Author information
Authors and Affiliations
Contributions
K.R., H.A., T.H. and Y.H. conceived and designed the experiments. K.R., T.G.Y. and Y.C. performed experiments. K.R., T.G.Y., H.A. and Y.H. analyzed the data. H.K., H.M. and T.H. contributed reagents/materials/analysis tools. K.R., T.G.Y., H.A. and Y.H. wrote the paper with the help of H.K., Y.C., H.M. and T.H.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ruan, K., Yamamoto, T., Asakawa, H. et al. Histone H4 acetylation required for chromatin decompaction during DNA replication. Sci Rep 5, 12720 (2015). https://doi.org/10.1038/srep12720
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep12720
This article is cited by
-
Chromatin-based DNA replication initiation regulation in eukaryotes
Genome Instability & Disease (2023)
-
Aberrant CREB1 activation in prostate cancer disrupts normal prostate luminal cell differentiation
Oncogene (2021)
-
Su(Hw) primes 66D and 7F Drosophila chorion genes loci for amplification through chromatin decondensation
Scientific Reports (2021)
-
Effect of histone H4 tail on nucleosome stability and internucleosomal interactions
Scientific Reports (2021)
-
Chromosome-associated RNA–protein complexes promote pairing of homologous chromosomes during meiosis in Schizosaccharomyces pombe
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.