Abstract
Live poultry markets are a source of human infection with avian influenza A (H7N9) virus. On February 21, 2014, a poultry farmer infected with H7N9 virus was identified in Jilin, China and H7N9 and H9N2 viruses were isolated from the patient's farm. Reassortment between these subtype viruses generated five genotypes, one of which caused the human infection. The date of H7N9 virus introduction to the farm is estimated to be between August 21, 2013 (95% confidence interval [CI] June 6, 2013-October 6, 2013) and September 25, 2013 (95% CI May 28, 2013-January 4, 2014), suggesting that the most likely source of virus introduction was the first batch of poultry purchased in August 2013. The reassortment event that led to the human virus may have occurred between January 2, 2014 (95% CI November 8, 2013-February 12, 2014) and February 12, 2014 (95% CI January 19, 2014-February 18, 2014). Our findings demonstrate that poultry farms could be a source of reassortment between H7N9 virus and H9N2 virus as well as human infection, which emphasizes the importance to public health of active avian influenza surveillance at poultry farms.
Similar content being viewed by others
Introduction
By June 27, 2014, 450 confirmed H7N9 human cases resulting in 165 deaths had been reported to the World Health Organization (WHO)1, following the identification of the first human case in Eastern China in March 20132. Most human infections are associated with a history of exposure to live poultry markets (LPMs)3,4,5,6,7 and the closure of LPMs is effective at controlling human infection8,9. No human H7N9 cases associated with poultry farms were reported prior to the identification of a poultry farmer infected with the H7N9 virus on February 21, 2014 in Jilin Province in northeastern China10. On February 19, 2014, throat swabs were collected from a 50-year-old male poultry farmer hospitalized in the Jilin University First Hospital and H7N9 virus was identified10 as A/Jilin/10117/2014 (H7N9) by the Chinese National Influenza Center (CNIC).
Avian influenza H7N9 virus is a triple-reassortant virus with a hemagglutinin (HA) gene derived from H7N3, a neuraminidase (NA) gene potentially from H7N9, H2N9 or H11N9 and six internal genes derived from H9N2 viruses2,4. It has been proposed that sequential and dynamic genetic reassortment of internal genes generates multiple genotypes of H7N9 viruses with better adaptation11,12,13,14; however, no actual reassortment events in poultry have been discovered to support this hypothesis. This study is the first to describe the reassortment process between H7N9 and H9N2 viruses co-circulating on a poultry farm and resulting in multiple genotypes, one of which was capable of causing human infection. Additionally, the upper and lower bounds of the introduction date of the H7N9 virus to the patient's farm and the upper bound of the reassortment event that led to the human H7N9 strain were estimated using the eight gene segments of the H7N9 and H9N2 viruses. Our findings indicate that poultry farms play an important role, similar to that of LPMs, in the genesis of novel H7N9 reassortments and could act as an important source of human infection.
Results
Field investigation identified H7N9 and H9N2 viruses on epidemiologically linked farms
Epidemiological investigations were initiated on February 20 to trace the source of H7N9 viruses causing human infection. The patient began to manage this small-scale poultry farm in August 2013 and approximately 600 chickens, 140 guinea fowl (Numidameleagris), 173 black-bone Silkie chickens (Gallus gallusdomesticus), 6 turkeys and a goose were raised together in a warehouse. Between August 2013 and February 2014, a total of seven batches of poultry were introduced to this farm (Fig. S1 in the Supplementary Information). In August 2013, 6 turkeys and 33 guinea fowl (N. meleagris) were introduced from an agricultural fair that was held August 16–25, 2013 in Changchun City. In October 2013, approximately 107 guinea fowl (N. meleagris) and 173 black-bone Silkie chickens (G. gallusdomesticus) were introduced from a farm in Siping City, some of which subsequently sickened and died. More black-bone Silkie chickens than the other types of poultry died. A local veterinarian reported that the cause of death of this batch of birds resulted from air sacculitis and enteritis. Testing for avian influenza virus was not performed. On December 24, 2013, 97 chickens were introduced from a dealer's farm that was sufficiently disinfected and no deaths occurred in this batch of birds. On December 27, 2013, the same dealer acquired 170 chickens from two free-range farmers in Yongji County and delivered those chickens to the patient's farm. Additionally, a poultry worker hired by the patient brought one goose and 4–5 chickens onto the farm. On February 3 and February 10, the second dealer introduced approximately 290 chickens from free-range farms in Xiaoyang in Dongfeng County onto the patient's farm. On February 12, the same dealer introduced the last batch of 90 chickens obtained in Wuxinghong in Dongfeng County to the patient's farm. Subsequent signs of illness and death widely occurred after the sixth batch of chickens were introduced on February 10 and approximately 200 chickens had died by February 22. The patient had checked the farm daily without personal protection and developed illness on February 15. No close contacts presented clinical symptoms, except for one individual with fever who had accompanied the patient when consulting the veterinarian for a diagnosis of the dead chickens. A series of throat swabs collected from this close contact were negative for influenza A virus. Paired serum samples collected from close contacts were seronegative for H7N9 virus infection (Table S1).
We collected a total of 279 samples from four epidemiologically linked farms (Table 1) as follows: 38 poultry samples and 46 environmental samples from the patient's farm (Site 1); 15 environmental samples from the first dealer's farm and the backyards of the free-range farmers from whom chickens had been obtained and introduced between December 24 and December 27, 2013 (Site 2); 12 poultry samples and 153 environmental samples from the backyards of the free-range farmers in Dongfeng County from whom the second dealer had obtained chickens for the last three batches in February (Site 3); and 6 poultry samples and 9 environmental samples from the backyards of the close contact who developed a fever (Site 4). The viruses were isolated only from the patient's farm and they included six H7N9 viruses from oropharyngeal or cloacal swabs of chickens and three H9N2 viruses from oropharyngeal swabs of chickens and drinking water.
Molecular markers associated with mammalian adaptation and drug resistance were identified in the H7N9 viruses
Selected amino acid substitutions associated with mammalian adaptation, viral replication and drug resistance were analyzed (Table S2). All seven H7N9 viruses, including the human isolate, demonstrated the G186 V mutation in the HA gene and the Q226 L mutation, with the exception of one virus isolated from a chicken that contained 226Q; these results indicated an increased preference for human-like receptor (α2–6) binding15,16. All of the NA genes examined possessed a 69–73 amino acid deletion in the stalk region, which is related to enhanced virulence in mice as well as adaption and transmission in domestic poultry17. Although the viruses isolated from the farm contained 701D and 627E, the human isolate, A/Jilin/10117/2014 (H7N9), possessed a D701N mutation without an E627K mutation in the polymerase basic protein 2 (PB2) gene, which has been proposed to be associated with high polymerase activity and increased virulence in mice18,19. The PB1 L368V mutation, which increases transmission in ferrets20, was identified in all of the viruses. The signature amino acids of the human influenza virus polymerase acidic protein (PA) gene were detected, including K356R and S409N21. Drug resistance mutations were detected in the matrix protein 2 (M2) gene (S31N); however, the E119V/A/D, R152K, H274Y and R292K mutations were not detected in the NA gene, indicating that the H7N9 viruses were resistant to amantadine and sensitive to NA inhibitors22,23,24.
Multiple genotypes of H7N9 were generated as a result of co-circulation of H7N9 and H9N2 viruses on the poultry farm
Phylogenetic trees showed that the H7 and N9 genes of the human isolate clustered with the novel H7N9 viruses in the public sequence databases (Fig. S2 in the Supplementary Information). The nucleoprotein (NP), non-structural (NS) and PB1 genes were identified within the major clade, whereas the PB2 and matrix protein (MP) genes fell within the minor clade of the previously reported H7N9 viruses (Fig. S3 in the Supplementary Information). Unlike other segments, the PA gene was different from the H7N9 viruses reported previously and clustered with the H9N2 viruses isolated from the patient's poultry farm (Fig. 1).
Phylogenetic tree of the PA gene.
The branches in orange represent the H7N9 viruses and those in black represent the H9N2 viruses. The branches in red represent the H7N9 and H9N2 viruses isolated in this study. The names of the viruses are labeled in the adjacent square frames. The red branches represent the H7N9 viruses and those in blue represent H9N2 viruses.
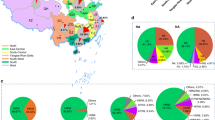
With regard to the six internal genes of all the H7N9 and H9N2 isolates, each gene formed two groups (A and B) on the phylogenetic trees (Fig. S3 in the Supplementary Information). The sequence similarity between the groups of each gene was lower than 99.6% (Fig. 2a and Tables S5–S10). Seven genotypes were designated, based on the combination of different groups of individual internal genes (Fig. 2c). The genotype with the combination of “AABBAA,” as represented by the A/chicken/Jilin/13188/2014 (H7N9)-like virus (Ck13188-like) and a genotype of “BBAABB,” as represented by the A/chicken/Jilin/13200/2014(H9N2)-like virus (Ck13200-like), provided internal genes as the parental viruses to the other genotypes, including the genotype that caused the human infection (Fig. 2c). A/Chicken/Jilin/13204/2014(H9N2) and A/Chicken/Jilin/13199/2014(H7N9) represent combinations containing two genotypes (Fig. 2b, c). The likely scenario for the reassortment between the H7N9 and H9N2 viruses co-circulating on the poultry farm is illustrated in Fig. 3.
The percentage of identity for the six internal genes and the divergent viral genotypes isolated in this study.
(a) The identity of the six internal genes of the seven H7N9 viruses and three H9N2 viruses isolated in this study. Each gene formed two groups, assigned as group A and B, based on a sequence similarity higher than 99.6%; those two groups are shown in green. The “mix” marked in the square of the PB2 and MP genes represents a mixture of the amino acids observed in the virus. (b) The mixed amino acids sites of the PB2 gene of virus A/Chicken/Jilin/13204/2014(H9N2) and the MP gene of virus A/Chicken/Jilin/13199/2014(H7N9) are specified. (c) A and B indicates group A and B, respectively and the different combination of groups in each segment represents different viral genotypes. Orange and green indicate the gene segments of the parental H7N9 and H9N2 viruses, respectively.
The reassortment scenario between H7N9 and H9N2 viruses co-circulating on the poultry farm.
The different species of poultry involved in the reassortment are shown in cartoons. The cartoon was created by BT and LY using Adobe Illustrator CS6. The virus is demonstrated with a circle. The eight gene segments (horizontal bars), from top to bottom, include PB2, PB1, PA, HA, NP, NA, MP and NS. Orange represents the genes from the parental H7N9 viruses. Green represents the genes from the parental H9N2 virus. The green and orange internal gene names in the frame represent the gene pools on the patient's farm.
The estimated date of the introduction and reassortment of H7N9 viruses
We determined the most recent common ancestors (TMRCAs) and 95% CI for 3 key nodes for each segment using the Bayesian Markov chain Monte Carlo approach in BEAST v1.825 as follows: 1) the TMRCA of the viruses isolated from the patient and his farm; 2) the TMRCA of the farm viruses plus the closest viruses isolated elsewhere; and 3) the TMRCA of the human virus and its closest avian virus on the farm. Items (1) and (2) provided the upper and lower bounds of the date of introduction of H7N9 to the farm, respectively and item (3) provided an upper bound for the date of the occurrence of the reassortment event that led to the human strain. According to our results, as shown in Fig. 4 and Fig. S4 in the Supplementary Information, the TMRCAs of the eight gene segments from item (1) ranged from September 25, 2013 (95% CI May 28, 2013-January 4, 2014) to January 18, 2014 (95% CI, November 7, 2013-February 7, 2014); the M gene of group A was the earliest gene identified with the TMRCA on September 25, 2013 (95% CI May 28, 2013-January 4, 2014). The TMRCAs of the eight segments from item (2) ranged from April 18, 2010 (95% CI June 26, 2009-November 25, 2010) to August 21, 2013 (95% CI June 6, 2013-October 6, 2013); the N9 gene was the latest gene identified with the TMRCA on August 21, 2013 (95% CI June 6, 2013-October 6, 2013). These results indicated that H7N9 might have been introduced to the patient's farm between August 21, 2013 (95% CI June 6, 2013-October 6, 2013) and September 25, 2013 (95% CI May 28, 2013-January 4, 2014). Considering the dates of the introduction of the seven batches of poultry, the poultry batch acquired in August 2013 represents the likely source of H7N9 introduced to the farm. According to the TMRCA of H7N9 from item (3), the reassortment event that led to the human strain might have occurred between January 2, 2014 (95% CI November 8, 2013-February 12, 2014) and February 12, 2014 (95% CI January 19, 2014-February 18, 2014).
The estimated date of the introduction of H7N9 onto the patient's farm and the reassortment event that led to human infection.
The red circles and blue bars represent the estimated TMRCA and 95% CI, respectively, of each gene segment of the viruses isolated from the patient's farm; the black circles and orange bars represent the TMRCA and 95% CI, respectively, of each gene segment of the viruses isolated from the farm and the closest relatives sampled elsewhere; the black triangles and dotted lines represent the TMRCA of each gene segment of the human virus and the closest avian viruses isolated from the farm. The black arrows pointing to the grey line represent the dates of the seven batches of introduced poultry. The green column represents the upper and lower bounds of the introduction date of H7N9 onto the farm. The magenta column represents the upper and lower bounds of the reassortment event that led to the human strain occurrence.
Discussion
The novel avian influenza A (H7N9) virus is a triple reassortant of H7N3, H7N9 (H2N9, H11N9) and H9N2 viruses with low pathogenicity in poultry2,26,27; however, this virus causes severe illness in humans, including rapidly progressive pneumonia, respiratory failure, acute respiratory distress syndrome (ARDS), high rates of intensive care unit (ICU) admission and fatal outcomes28,29. LPM exposure plays a major role in the human risk of H7N9 infections and LPM closures have reduced the incidence of human infection with the H7N9 virus9. Traces of H7N9 were identified recently at a chicken farm in Guangdong Province that supplied poultry to Macau, China30. However, no human cases associated with poultry farms were reported before the first patient infected with H7N9 was identified in Jilin in northeastern China10.
Many studies have suggested that H7N9 underwent dynamic genetic reassortment with H9N2 viruses to generate diversified genotypes12,13,14,31. However, the process of generating diversified genotypes has not been observed. In this study, reassortment between H7N9 and H9N2 viruses co-circulating on the same farm was documented (Fig. 3). Based on the phylogenetic tree and the similarity analysis of six internal genes, the parental Ck13188-like H7N9 and Ck13200-like H9N2 viruses entered the farm and generated four novel genotypes of H7N9 and one genotype of H9N2, with one of the H7N9 genotypes causing human infection. The unique PA gene of the human isolate was different from the genes of previously reported H7N9 viruses; however, the gene was similar to those of the H9N2 viruses isolated from the patient's farm, further supporting the hypothesis that the virus was generated by reassortment between H7N9 and H9N2 coexisting on the farm. No H7N9 viruses had been reported in Jilin prior to the identification of this human infection. The introduction of H7N9 viruses likely occurred between August 21, 2013 (95% CI June 6, 2013-October 6, 2013) and September 25, 2013 (95% CI May 28, 2013-January 4, 2014). According to the epidemiology data, seven batches of poultry were introduced onto the patient's farm and the virus most likely entered the farm from the first batch, which included 6 turkeys and 33 guinea fowl (N. meleagris) purchased from an agricultural fair in Changchun City in August 2013. Furthermore, the lack of reports showing H7N9 viruses on other epidemiologically linked farms further supports our conclusion of genetic reassortment.
The continued introduction of different species of poultry and the mixed breeding model may have contributed to the diversified genotypes generated by co-circulation of the H7N9 and H9N2 viruses on the patient's farm. Nearly 1,000 birds of five species including chickens, guinea fowl, black-bone Silkie chickens, turkeys and geese were involved in this process. Chickens were the most prevalent poultry species on the patient's farm. Previous studies have suggested that H7N9 and H9N2 predominately circulate in chickens27,32, indicating that the genesis of the diversified genotypes most likely occurred in chickens. However, the mixed breeding model may also facilitate the transmission of H7N9 and H9N2 among different species of poultry and expand the host range. Thus, species other than chickens might contribute to the genesis of different genotypes and human infection on poultry farms.
The H7N9 virus HA protein shows a binding affinity for mammalian receptors; however, no evidence of sustained human-to-human transmission has been reported. Similar to the findings of previous studies, clinical syndromes were not detected in any close contacts of the patient, except in an individual who developed a fever; influenza A was not detected in this contact. Moreover, traces of H7N9 viruses were not detected in the poultry or environmental samples collected from the backyards of the close contact and no H7N9 antibody was detected in the paired sera of any of the contacts.
There are some limitations to our study. For instance, a genotype identical to the human isolate was not detected in poultry, which may have been caused by the insufficient sample size. Nevertheless, according to the homology analysis, each gene with high identity (99.9%–100%) to A/Jilin/10117/2014 (H7N9) was detected in the viruses isolated from the poultry farm (Tables S3–S10). In addition, we proposed that the possible original source of the introduction of H7N9 was from the first batch of poultry purchased from the agricultural fair in August, 2013; however, without collecting samples from this batch of poultry, the actual original source of H7N9 virus could not be confirmed.
Our findings imply that poultry farms may act as a source for the generation of divergent genotypes of H7N9 viruses and human infection. Extensive poultry farm surveillance is therefore needed for public health security.
Methods
Ethics statement
As a public health response to the outbreak, written informed consent of the study subjects could be waived according to Chinese law. However, we obtained the patient's written informed consent for publication. The clinical sample collection and transportation were performed according to the Chinese Guidelines for the Diagnosis and Treatment of Human Infection with H7N9 Avian Influenza Virus (2nd edition, 2013). The experimental protocols were approved by the Laboratory Management Committee of the National Institute for Viral Disease Control and Prevention.
Surveillance, reporting and data collection
A standardized surveillance and reporting form was used to collect the epidemiological and clinical data, as follows: demographic characteristics; underlying medical conditions; history of seasonal influenza vaccination; recent exposure to poultry, swine or other animals; recent visits to a live animal market; clinical signs and symptoms; antiviral treatment; clinical complications; and outcomes. Active surveillance was performed by sampling the poultry and the environment of the patient's poultry farm and other possible sources of virus introduction. After identifying the close contacts of the patient, the medical conditions were monitored daily for 7 days after the last known exposure to the patient. Throat swabs were collected from the close contacts who developed clinical symptoms. Serum samples were collected in the acute and convalescent phases from the close contacts of the patient.
RNA extraction and real-time RT-PCR
Viral RNA was extracted using a QIAamp® Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Specific real-time reverse-transcriptase-polymerase-chain-reaction (RT-PCR) assays for seasonal influenza viruses (H1, H3 or B) and avian influenza H1 to the H16 and N1 to the N9 subtypes were performed to verify the viral subtypes.
Hemagglutination-inhibition (HI) assay
An improved horse erythrocyte HI assay with a modified order of serum treatment procedures was used to detect antibody response to H7N933,34. Beta-propriolactone (BPL)-inactivated reference H7N9 virus A/Anhui/1/2013 was used as the antigen in this assay.
Virus isolation and genome sequencing
The specimens were maintained in a viral transport medium and were inoculated and grown in 9- to 10-day-old specific pathogen-free (SPF) embryonated chicken eggs for 48–72 h at 37°C. The full genomes of the virus isolates were amplified with a QiagenOneStep RT-PCR Kit for sequencing (Qiagen, Germany). The PCR products were purified from an agarose gel with a QIAquick Gel Extraction Kit (Qiagen, Germany). Sanger sequencing was performed with an automatic Applied Biosystems 3730xl DNA Analyzer (Life Technologies, USA) and an Applied Biosystems Big Dye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, USA) according to the manufacturer's instructions. Deep sequencing was performed using the Ion torrent PGM platform. The library of 200-bp fragments was sequenced with a 318v2 chip. The next-generation sequencing (NGS) reads were analyzed with a pipeline process using the CLC Genomics workbench 7.0.4. The human isolate A/Jilin/10117/2014 and A/Chicken/Jilin/05/2014 were sequenced by Sanger sequencing. The sequences of other viruses isolated from chickens and the poultry farm environment were examined using deep sequencing with the Ion torrent PGM platform to investigate whether potential avian influenza viruses coexisted in the hosts. The accession codes of the viral sequences isolated in this study are shown in Table S11.
Genetic analysis
The percentage of each viral gene that was identified was calculated using MegAlign7.1. The datasets were obtained from the Global Initiative on Sharing All Influenza Data (GISAID) EpiFlu™ Database. A maximum likelihood phylogenetic tree for the nucleotide sequences of each gene of the selected influenza viruses was constructed with MEGA5.1. The bootstrap method was used to test the phylogeny and was run up to 1,000 times. The time in generations to TMRCA for the eight gene fragments was estimated with the Bayesian Markov chain Monte Carlo (MCMC) approach in BEAST v1.825 using the SRD06 nucleotide substitution model35. The uncorrelated lognormal relaxed clock model was selected for the rate variation among the branches. A Bayesian skyride was used as a prior coalescent tree and the UPGMA tree was used as the starting tree. For the prior parameter, ucld.mean and ucld.stdev were set as the uniform distributions; the default values of the other parameters were used. The virus isolates without a detailed sampling date were assigned to the middle of the year/month. The MCMC algorithm was run for 5 × 107 steps for each dataset, with sampling every 500 steps. Convergence of the parameters was determined as effective sample sizes (ESSs) larger than 200 using Tracer v1.6. The maximum clade credibility trees with median node heights are summarized.
References
World Health Organization (WHO). WHO Risk Assessment as of 27 June 2014. (2014). (http://www.who.int/influenza/human_animal_interface/influenza_h7n9/riskassessment_h7n9_27june14.pdf)(Accessed 27 June 2014).
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368, 1888–1897, 10.1056/NEJMoa1304459 (2013).
Wang, C. et al. Relationship between domestic and wild birds in live poultry market and a novel human H7N9 virus in China. J Infect Dis 209, 34–37, 10.1093/infdis/jit478 (2014).
Chen, Y. et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–25, 10.1016/S0140-6736(13)60903-4 (2013).
Bao, C. J. et al. Live-animal markets and influenza A (H7N9) virus infection. N Engl J Med 368, 2337–2339, 10.1056/NEJMc1306100 (2013).
Li, J. et al. Environmental connections of novel avian-origin H7N9 influenza virus infection and virus adaptation to the human. Science China. Life sciences 56, 485–492, 10.1007/s11427-013-4491-3 (2013).
Shi, J. D. G. & Liu, P. Isolation and characterization of H7N9 viruses from live poultry markets—Implication of the source of current H7N9 infection in humans. Chinese Sci Bull 16, 1857–1863, 10.1007/s11434-013-5873-4 (2013).
Cowling, B. J. et al. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382, 129–37, 10.1016/S0140-6736(13)61171-X (2013).
Yu, H. et al. Effect of closure of live poultry markets on poultry-to-person transmission of avian influenza A H7N9 virus: an ecological study. Lancet 383, 541–548, 10.1016/S0140-6736(13)61904-2 (2014).
Fan, M. et al. Human Influenza A(H7N9) Virus Infection Associated with Poultry Farm, Northeastern China. (2014). (http://www.nc.cdc.gov/eid/article/20/11/14-0608_article)(Accessed 17 July 2014).
Liu, D. et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural and coalescent analyses. Lancet 381, 1926–3, 10.1016/S0140-6736(13)60938-1 (2013).
Cui, L. et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun 5, 3142, 10.1038/ncomms4142 (2014).
Wu, A. et al. Sequential reassortments underlie diverse influenza H7N9 genotypes in China. Cell Host Microbe 14, 446–452, 10.1016/j.chom.2013.09.001 (2013).
Lu, J. et al. Continuing reassortment leads to the genetic diversity of Influenza H7N9 in Guangdong China. J Virol 88, 8297–306. 10.1128/JVI.00630-14. (2014).
Dortmans, J. C. et al. Adaptation of novel H7N9 influenza A virus to human receptors. Sci Rep 3, 3058, 10.1038/srep03058 (2013).
Zhou, J. et al. Biological features of novel avian influenza A (H7N9) virus. Nature 499, 500–503, 10.1038/nature12379 (2013).
Wang, Q. Z. et al. Biological significance of amino acids deletion in NA stalk of H5N1 avian influenza virus. Wei Sheng Wu Xue Bao 46, 542–546 (2006).
Mok, C. K. et al. Amino Acid Substitutions in Polymerase Basic Protein 2 Gene Contribute to the Pathogenicity of the Novel A/H7N9 Influenza Virus in Mammalian Hosts. J Virol 88, 3568–3576, 10.1128/JVI.02740-13 (2014).
Zhang, H. et al. The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J Gen Virol 95, 779–86, 10.1099/vir.0.061721-0 (2014).
Herfst, S. et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541, 10.1126/science.1213362 (2012).
Taubenberger, J. K. et al. Characterization of the 1918 influenza virus polymerase genes. Nature 437, 889–893, 10.1038/nature04230 (2005).
Sleeman, K. et al. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis 19, 1521–1524, 10.3201/eid1909.130724 (2013).
Woods, C. J., Malaisree, M., Long, B., McIntosh-Smith, S. & Mulholland, A. J. Computational assay of H7N9 influenza neuraminidase reveals R292K mutation reduces drug binding affinity. Sci Rep 3, 3561, 10.1038/srep03561 (2013).
Wang, D. et al. Genetic tuning of the novel avian influenza A(H7N9) virus during interspecies transmission, China, 2013. (2014). (http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20836)(Accessed 26 June 2014).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29, 1969–1973, 10.1093/molbev/mss075 (2012).
Lam, T. T. et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502, 241–244, 10.1038/nature12515 (2013).
Pantin-Jackwood, M. J. et al. Role of poultry in spread of novel H7N9 influenza virus in China. J Virol 88, 5381, 10.1128/JVI.03689-13 (2014).
Hai-Nv Gao, M. D. et al. Clinical Findings in 111 Cases of Influenza A (H7N9) Virus Infection. N Engl J Med 368, 2277–85, 10.1056/NEJMoa1305584 (2013).
Li, Q. et al. Preliminary Report: Epidemiology of the Avian Influenza A (H7N9) Outbreak in China. N Engl J Med 370, 520–32, 10.1056/NEJMoa1304617 (2013).
China: Guangdong farm culls chickens after H7N9 found in Macau. (2014). (http://crofsblogs.typepad.com/h5n1/2014/03/china-guangdong-farm-culls-chickens-after-h7n9-found-in-macau.html) (Accessed 17 March 2014).
Zhang, L., Zhang, Z. & Weng, Z. Rapid reassortment of internal genes in avian influenza A(H7N9) virus. Clin Infect Dis 57, 1059–1061, 10.1093/cid/cit414 (2013).
Kahn, R. E. & Richt, J. A. The novel H7N9 influenza A virus: its present impact and indeterminate future. Vector Borne Zoonotic Dis 13, 347–348, 10.1089/vbz.2013.999.ceezad (2013).
World Health Organization (WHO). Serological detection of avian influenza A(H7N9) infections by horse red blood cells haemagglutination-inhibition assay. (2013). (http://www.who.int/influenza/gisrs_laboratory/cnic_serological_diagnosis_hai_a_h7n9_20131220.pdf) (Accessed20 December 2013).
Dong, L. et al. A Combination of Serological Assays to Detect Human Antibodies to the Avian Influenza A H7N9 Virus. PLoS One 9, e95612, 10.1371/journal.pone.0095612 (2014).
Shapiro, B., Rambaut, A. & Drummond, A. J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 23, 7–9, 10.1093/molbev/msj021 (2006).
Acknowledgements
This study was supported by the National Mega-Projects for Infectious Diseases (2014ZX10004002 to Y Shu, 2013ZX10004-101 to DL, 2013ZX10004611-003 to JZ and 2012ZX10004215 partly to DW) and the Chinese National Influenza Center–Centers for Disease Control and Prevention (CDC) collaborative project 5U51IP000334-03 and the CDC China–US Collaborative Program on Emerging and Re-emerging Infectious Diseases.
Author information
Authors and Affiliations
Contributions
D.W., M.F. and Y.S. designed the study. Z.S., T.B., J.L., X.Z., H.L., X.L., X.Y., L.X., S.X., X.Z., X.L., A.W., J.G., B.S., W.H., Y.Z., X.L., R.G., B.S., J.D. and H.W. performed the field and laboratory experiments. L.Y., C.T. and D.W. analyzed and interpreted the data. S.W., Q.L., D.L., G.W., Z.F., G.F.G. and Y.W. commented on this study. T.B., Y.S., D.W. and L.Y. wrote the manuscript. T.B. created the cartoons of poultry shown in Figure 3. L.Y. and T.B. constructed Figures 1–4.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Wu, D., Zou, S., Bai, T. et al. Poultry farms as a source of avian influenza A (H7N9) virus reassortment and human infection. Sci Rep 5, 7630 (2015). https://doi.org/10.1038/srep07630
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07630
This article is cited by
-
The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status
Military Medical Research (2020)
-
A ten-year China-US laboratory collaboration: improving response to influenza threats in China and the world, 2004–2014
BMC Public Health (2019)
-
Attenuation of highly pathogenic avian influenza A(H5N1) viruses in Indonesia following the reassortment and acquisition of genes from low pathogenicity avian influenza A virus progenitors
Emerging Microbes & Infections (2018)
-
Multiple polymerase gene mutations for human adaptation occurring in Asian H5N1 influenza virus clinical isolates
Scientific Reports (2018)
-
Avian influenza A/H7N9 risk perception, information trust and adoption of protective behaviours among poultry farmers in Jiangsu Province, China
BMC Public Health (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.