Abstract
The molecular genetic mechanisms underlying abaxial-adaxial polarity in plants have been studied as a property of lateral and flattened organs, such as leaves. In leaves, laminar expansion occurs as a result of balanced abaxial-adaxial gene expression. Over- or under- expression of either abaxializing or adaxializing genes inhibits laminar growth, resulting in a mutant radialized phenotype. Here, we show that co-option of the abaxial-adaxial polarity gene network plays a role in the evolution of stamen filament morphology in angiosperms. RNA-Seq data from species bearing laminar (flattened) or radial (cylindrical) filaments demonstrates that species with laminar filaments exhibit balanced expression of abaxial-adaxial (ab-ad) genes, while overexpression of a YABBY gene is found in species with radial filaments. This result suggests that unbalanced expression of ab-ad genes results in inhibition of laminar outgrowth, leading to a radially symmetric structure as found in many angiosperm filaments. We anticipate that co-option of the polarity gene network is a fundamental mechanism shaping many aspects of plant morphology during angiosperm evolution.
Similar content being viewed by others
Introduction
The homology of floral organs to vegetative leaves has long been proposed as a fundamental principle of plant comparative morphology. Since 1790, Goethe1 envisioned what in more recent years has been described as ‘serial homology’ between vegetative and reproductive organs. For Goethe1, leaves and floral organs shared an underlying common theme that would manifest itself differently during specific phases of plant development, leading to a series of successive forms: leaves, bracts, sepals, petals, stamens (microsporophylls) and carpels (megasporophylls). More recently, the genetic mechanisms underlying plant organogenesis and specifically leaf and floral organ initiation, growth and development have been revealed2,3,4. Indeed, many of the same mechanisms that operate in shaping leaves seem to play an important role in shaping floral organs, with perhaps the exception of the mega- and microsporangia5. For instance, polarity genes, such as those described in leaf abaxial-adaxial patterning, have also been implicated in establishing abaxial-adaxial polarity of various floral organs (e.g. polarity of sepals and petals6,7,8; polarity in stamens and ovules9; polarity during ovule development10,11,12,13).
Abaxial-adaxial (ab-ad) polarity in plants has been most widely studied as a property of flattened lateral organs (i.e. the leaf). The first ab-ad polarity genes were described in Antirrhinum majus and Arabidopsis thaliana mutants with radially symmetric, or filamentous, leaves in place of the WT laminar structures6,14,15. Since then, much has been discovered about ab-ad polarity patterning and the underlying gene regulatory network responsible for the establishment of organ polarity, including the fundamental role of smallRNAs, such as tasiR-ARF and miR165/16616. Most importantly, it has been shown that laminar development of the type observed in leaves and many floral organs is dependent on a balanced expression of abaxial and adaxial identity genes. Disruption of either the abaxial or adaxial signaling pathway tends to produce a mutant phenotype of ‘radialized’ or radially symmetric leaves, with the entire leaf epidermis displaying markers characteristic of the adaxial surface in the case of abaxial signaling mutants, or the abaxial surface in the case of adaxial signaling mutants6,14,15.
The molecular processes implicated in laminar expansion in most model plants studied thus far is referred to as the ‘abaxial–adaxial juxtaposition hypothesis.’15 In this model, the location of laminar expansion is described as the contact zone of abaxializing (e.g., YABBY genes, KANADI genes) and adaxializing (e.g., REVOLUTA, PHABULOSA) gene products, where laminar outgrowth is initiated at and restricted to this contact zone. Unlike in Arabidopsis, however, YABBY gene expression in Zea mays is localized to the adaxial side of incipient and young leaf primordia17 and it has been suggested that this polarized expression of the YABBY genes may play a more direct role in mediolateral organ outgrowth rather than in cell fate determination per se17. Such results suggest that two underlying processes are co-occurring and mediated by recognized adaxializing and abaxializing genes: cell fate determination and mediolateral organ outgrowth. Regardless of their roles in cell fate determination, it is interesting to note that down-regulation or up-regulation of either abaxial or adaxial genes leads to an impairment of laminar growth, suggesting that a proper balance between abaxializing and adaxializing gene products is essential for laminar expansion. Despite the relatively detailed knowledge of the gene regulatory networks involved in determining the polarity of leaves and floral organs in model systems, we still know relatively little about how such networks have changed though evolutionary time or how they may be responsible for different leaf and floral morphologies observed during angiosperm evolution.
Here we present evidence for the involvement of polarity genes in the evolution of stamen morphological diversity through the study of the comparative development of androecial form across the Zingiberales. The evolution of floral morphology in this order is characterized by increased petaloidy of the androecial whorl organs, which is inversely correlated to the number of fertile stamens produced by each flower (Figure 1). The evolution of petaloid, laminar stamen filaments from an ancestral radially symmetric filament and the development of both sterile and fertile petaloid stamens within the Zingiberales provide unique opportunities for the study of the role of polarity genes in shaping filament morphology (Figure 2). We hypothesize that the laminar expansion occurring in the filaments of the derived species in the order is a result of altered expression of polarity genes as compared with polarity gene expression in species with ancestral, radial filaments. According to this hypothesis, the gene expression pattern observed in petaloid stamens restores the balance between abaxial and adaxial gene products within the stamen primordium, leading to the elaboration of a margin that functions to signal laminar expansion in an otherwise radialized primordium. In order to test this hypothesis, we generated a set of organ-specific RNA-Seq data for floral organs of Musa basjoo and Costus spicatus, including radial (Musa) and petaloid (Costus) stamen filaments. The results were compared to gene expression in the radial filament of Brassica rapa B3 (Edger et al. unpublished) using the qTeller public database (Schnable and Freeling 2013, http://qteller.com/) and confirmed by qPCR (Supplement Figure 1).
Phylogenetic relationships of and androecial morphological evolution within the Zingiberales order.
This order of herbaceous tropical monocots of approximately 2,500 species comprises eight families which are generally divided into two groups: the basal paraphyletic banana group - including Musaceae, Strelitziaceae, Lowiaceae and Heliconiaceae - and the ginger clade comprising Zingiberaceae, Costaceae, Marantaceae and Cannaceae31. The arrow points to an important transition in the evolution of androecial morphology in the Zingiberales; at the base of the ginger clade, there is an impressive reduction in the number of fertile stamens, from 5 or 6 in the banana group to 1 or ½ in the ginger clade. This reduction in the number of fertile stamens is also associated with increased petaloidy of the infertile androecial members (i.e., staminodes). Furthermore, fertile stamen members in the ginger clade families, such as Costaceae and Cannaceae also show laminar expansion of the filament30, leading to a petaloid fertile stamen. Photos depict the fertile filament of Canna indica (Cannaceae) at the top; Costus spicatus (Costaceae) in the middle; and Musa basjoo (Musaceae) at the bottom right. th – thecae; fil – filament. The * marks the transition between the theca and the filament within the banana stamen.
Musa and Costus flowers, stamen filaments/connective and vasculature.
Schematic diagram of Costus and Musa inflorescences and position of the flowers with respect to the inflorescence meristem. Costus flowers are arranged in a spiral manner with younger flowers toward the top, while Musa flowers are arranged in two arrays (hands) subtended by a large bract. Usually, Musa inflorescences start the development of female flowers and as development proceeds, male flowers are produced later on. Costus spicatus single fertile stamen has a large laminar filament and connective and the filament, in particular, tends to expand later in development. All other androecial members are infertile and petaloid in nature (labellum). Musa basjoo five fertile stamens have a typical quasi-radial short filament bearing extensive theca. In Costus, filament vasculature shows an abaxial-adaxial distribution of phloem-xylem, similar to what is expected for other laminar structures such as leaves. In Musa, however, the organization of vascular bundles is quite unique, with two groups of xylem cells adjacent to phloem cells in the median plan. Thus, in Musa, filament vasculature shows a lateral-medial polarity axis, instead of a abaxial-adaxial, or a radially symmetrical pattern. Drawings made by AMRA.
Results
Based on the results of a reciprocal BLAST search, probable orthologs of Arabidopsis thaliana ab-ad polarity genes18 were identified from Musa acuminata and Costus spicatus (Table 1). In many cases, there was a one-to-many, or a many-to-one relationship between Arabidopsis genes and either Musa or Costus genes. Only genes that were present in Costus spicatus and Musa basjoo transcriptomes were used for downstream expression analyses. With only few exceptions, BLAST results identified a one-to-one relationship between Musa acuminata and Costus spicatus genes, which facilitated downstream expression analyses.
Starting with twelve characterized Arabidopsis thaliana ab-ad polarity genes, twenty-one presumed orthologs were identified within the Musa acuminata genes and fifteen orthologous genes were retrieved from the Costus spicatus transcriptome assembly, in line with the lineage-specific whole genome duplication proposed for Musaceae by the Musa acuminata genome sequencing group19.
FPKMs were calculated for the recovered abaxial-adaxial polarity genes using eXpress. Transcriptome replicates for all sampled floral organs resulted in similar FPKM counts for polarity genes in both Costus spicatus and Musa basjoo replicates. A mean FPKM was thus calculated across both replicates (Table 1).
Overall, ab-ad polarity genes in the filament of Musa basjoo and Costus spicatus showed low expression values in the RNA-seq datasets. The exception was MusaGSMUA_Achr7G01330_001, a YABBY2/5 ortholog (Figure 3a). In Musa basjoo filaments, this gene showed a 25-fold increase in FPKM when compared to Costus spicatus filament expression (15.7 in Costus; 395.8 in Musa). The orthologous gene from the Costus spicatus transcriptome, YABBY2/5 orthologue comp35601_c0_seq1, showed a low expression in the filament, equivalent to those found for other polarity genes in the filament. Interestingly, the expression of MusaGSMUA_Achr7G01330_001 is 4-fold higher in the filaments of Musa basjoo than in the free petal, a laminar structure within the Musa flower. When filament expression of the other YABBY2/5 orthologs was compared between Musa basjoo and Costus spicatus, no correlation was found between differential expression and differential morphology. For all other YABBY2/5 homologues, Musa and Costus filaments showed statistically similar FPKM values (Figure 3a).
Fragments Per Kilobase of exon per Million fragments mapped (FPKM) for Musa basjoo, Costus spicatus and Brassica rapa ab-ad polarity genes.
Musa acuminata genome was used as reference nomenclature for Musa basjoo and Costus spicatus gene expression. (a) Musa basjoo vs. Costus spicatus ab-ad polarity gene expression. Relative FPKM of all ab-ad polarity genes analyzed for Musa basjoo and Costus spicatus filaments are shown. Note that, overall, polarity genes in the filament show low expression values in both species, with the exception of one single gene in Musa basjoo (arrow), GSMUA_Achr7G01330_001. (b) Ab-ad polarity gene expression profiles (based on FPKM) of filaments from Musa basjoo, Costus spicatus and Brassica rapa. Brassica rapa shows a similar pattern of gene expression to that observed in Musa, where gene, in general, polarity genes have low FPKM values, except for a YABBY2/5 homologue. Mean FPKM values are presented based on two technical replicates for each tissue in each species.
In Brassica rapa, a distantly related species with radial filament morphology similar to that of Musa filaments, the expression of YABBY2 and YABBY5 genes show a similar pattern of expression. Brassica rapa has three YABBY2 and one YABBY5 homologues. In the Brassica rapa filament transcriptome, most genes, including YABBY genes, are expressed at very low levels. However, YABBY2-2A shows a 9-fold increase in FPKM when compared to expression of other YABBY2 or YABBY5 genes in the same organ (Figure 3b). Also, YABBY2-2A shows at least a two-fold increase in expression when compared to leaves or sepals of the same species. Although B. rapa petals show high expression levels of YABBY2-2A, the levels of expression of this gene are still significantly lower than those from filament transcriptomes (data not shown).
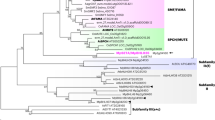
A phylogenetic analysis of YABBY genes across angiosperms was performed using maximum likelihood to confirm the evolutionary relationship of Musa and ArabidopsisYABBY genes. The gene tree (Figure 4) corroborates our BLAST results based on similarity. The MusaYABBY2/5 gene is resolved within the YABBY2 clade of Arabidopsis thaliana. It is worth noting that monocots do not have a representative within the ArabidopsisYABBY5 clade and that YABBY2 and YABBY5 clades are sister clades. Thus, we choose to call our gene a YABBY2/5 homologue, as the duplication resulting in the separate YABBY2 and YABBY5 gene lineages most likely occurred after the divergence of monocots and eudicots.
Maximum Likelihood analysis of YABBY genes across angiosperms.
Musa genes were retrieved from the banana genome19. Numbers on the branches represent bootstrap support for 100 replicates. Marked clades are based on Arabidopsis thaliana genes. Note that YABBY-2 and YABBY-5 genes are sister clades and monocots species only have representatives on the YABBY-2 clade. Arrow points to overexpressed Musa YABBY gene.
In order to confirm results obtained from RNA-Seq experiments, quantitative-PCR (qPCR) was performed with tissue from Musa acuminata filament and free petal and Costus spicatus labellum (fused petaloid staminodes) and laminar filament. Expression for YABBY2/5 ortholog (GSMUA_Achr7G01330_001, comp35601_c0_seq1); KANADI2/3 ortholog (GSMUA_Achr10G06430_001, comp30224_c0_seq1); and AS1 (GSMUA_Achr9G03320_001, comp38033_c0_seq1) shows concordant pattern between RNA-Seq and qPCR: low expression was detected for all genes in the filament when compared to YABBY2/5 expression levels in Musa acuminata (Figure 5). It is interesting to note that although RNA-Seq results indicate a potential role of ASSYMETRIC LEAF 1 (AS1) ortholog in the downregulation of YABBY2/5 in Costus filaments, quantitative PCR does not support this result, although it suggests a potential role of AS1 in the formation of the labellum. It might be the case that different regulatory mechanisms are operating in an organ-specific manner to down-regulate YABBY2/5 expression. A deeper understanding of YABBY2/5 downregulation and AS1 overexpression in Costus spicatus awaits further studies.
In situ hybridization experiments to detect gene expression during early development of Costus spicatus flowers show expression of YABBY2/5 in the adaxial side of the laminar androecial members (Figure 6a,b). Adaxial YABBY2/5 expression is more evident in the labellum, although it can also be observed in the C.spicatus fertile stamen filament/connective (Figure 6a, arrow). Environmental scan electron microscopy of abaxial and adaxial surfaces of Costus spicatus filament show similar cell types, with almost undistinguishable characteristics, suggesting that adaxial expression of YABBY2/5 does not lead to differences in cell type at the microscopic level.
In situs hybridization of Costus spicatus YABBY 2/5 genes and environmental electron microscopy of Costus spicatus filament abaxial and adaxial surfaces.
(a) Localization of YABBY 2/5 expression in the androecial members of Costus spicatus developing flower. Expression can be more clearly observed in the labellum (la). Arrow points to a patch of differential expression in the filament (fil). (b) Sense probe used as negative control. Environmental scan electron microscopy of abaxial (c) and adaxial (d) surfaces of Costus spicatus filament. se – sepal; pe – petal; la – labellum; fil – filament; th – theca; ct – connective; gy – gynoecium; fb – flower bract.
Discussion
The abaxial-adaxial polarity gene network has been independently implicated as a potential developmental mechanism for various morphological traits during land plant evolution, such as leaf lamina and ovule integument development11,13,18,20,21. Most of the genes that make up this network are shared by at least all seed plants (i.e. YABBY genes22) or even by all land plants (Streptophytes) (i.e., HD-ZIPIII genes23), suggesting that this network, or at least some of its components, is shared by all vascular plants.
In particular, the YABBY gene family has five distinct members in eudicots: FILAMENTOUS FLOWER (FIL), YABBY2, CRABS CLAW (CRC), INNER NO OUTER (INO) and YABBY522. Functional studies in Arabidopsis thaliana have supported the idea that some of the members of the YABBY family (more specifically FIL, YABBY2 and YABBY5) play important roles in both abaxial cell fate and abaxial/adaxial juxtaposition-mediated lamina expansion20 in angiosperm lateral organs, while others (particularly CRC and INO) are expressed specifically in the carpel and are important for ovule integument development10. Studies in maize, on the other hand, support the idea that YABBY genes are expressed on the adaxial side of the lateral organ primordium and are perhaps more directly involved in mediolateral (laminar) organ growth than cell fate determination, suggesting different evolutionary trajectories for the YABBY genes in monocot v. eudicot lineages17.
Recent studies on YABBY diversification in angiosperms suggest that abaxial expression of YABBY5 and FIL in lateral organs is an ancestral pattern, as is carpel expression of INO and CRC. However, restriction of CRC expression to the carpel was acquired later after the divergence of the Nymphaeales22. Thus, throughout the history of the vascular plants the leaf ab-ad polarity network has been co-opted for the specification of floral organ polarity, such as the case for ovule integument polarity. If we define co-option as the use of pre-existing traits – including genes and/or entire gene regulatory networks or modules – in a new way and presume that co-option can ultimately generate novelties24, we can argue that the involvement of the leaf ab-ad polarity gene regulatory network underlying androecial petaloidy in the Zingiberales is a clear case of co-option. Moreover, extrapolating from the data obtained from Brassica rapa filaments, ab-ad polarity genes seem to play a role in stamen morphology throughout the angiosperms and appear thus to form a fundamental network that regulates laminar growth across the entire plant body plan.
It is striking to notice that laminar, petaloid filaments and radially symmetric filaments have distinct macro and microscopical morphology (Figure 2). These morphological differences parallel those described for the controversial distinction between megaphyllous and microphyllous leaves25. It has been shown that at least some of the molecular mechanisms specifying the development of microphylls and megaphylls are shared between microphyll-bearing lycopodes and megaphyll-bearing euphyllophytes (ferns, gymnosperms and angiosperms), demonstrating the importance of consideration of physical constraints when comparing structures of differing size but similar position and/or function26. It is also interesting to notice that YABBY2/5 gene is expressed at the adaxial surface of the fertile stamen of C.spicatus flowers, in agreement to what has already been suggested for expression patterns in maize17. Moreover, scanning electron microscopy of Costus spicatus filaments shows similar cell types on both abaxial and adaxial surfaces of the fertile stamen, suggesting that differential expression of YABBY2/5 is not correlated with differential identity or cell differentiation patterns. As has already been suggested for maize17, our results suggest that YABBY2/5 in the Zingiberales is likely to be correlated to specification of laminar outgrowth rather than cell fate identity.
Studies on the molecular mechanisms of cladode development also demonstrate the role of YABBY genes and the ab-ad polarity network in the alteration of radial stem morphology to form a laminar, leaf-like stem ( = cladode) in Asparagales27,28. Whether of determinate (e.g. leaf and flower) or indeterminate (e.g. stem) growth, lateral organ morphology in vascular plants seems to be intrinsically correlated with position relative to the shoot apex and the proper function of the ab-ad polarity network promoting laminar expansion. In this broader sense, whether a lateral structure will develop into a radial or flattened organ is dependent on balanced expression of abaxializing and adaxializing gene products in the lateral organ. Unbalanced gene expression will result in the default state of radial growth. Thus, a single molecular mechanism appears to have been co-opted throughout the evolution of the angiosperms to shape lateral organ morphology, regardless of the identity of the lateral organ itself. Here, in line with previous studies of lateral organ development in angiosperms, we argue that the ab-ad polarity gene regulatory network is an ancestral developmental regulatory module guiding the development of flattened versus radial structures in angiosperm lateral organs, including the floral organs.
It is important to point out that the results presented here only focus upon those genes retrieved on our transcriptome analysis. It is possible, however, that we have not retrieved all the ab-ad polarity genes and other genes might be relevant for the evolution and development of the laminar filament in Costus. Also, it is likely that we have not retrieved all possible gene copies for all the ab-ad polarity gene families. A more extensive study for each of the polarity families could provide a broader understanding of the role of this network in the evolution of filament morphology in the Zingiberales.
Our results demonstrate the role of polarity genes in the evolution of stamen morphology and provide further support for the hypothesis that that the same underlying molecular mechanisms functioning in determining leaf shape and morphology have been co-opted during the evolution of angiosperms to effect organ morphology in flowers, specifically in the androecial whorl. Our results further suggest that the abaxial-adaxial polarity gene network is one of the fundamental modules involved in shaping plant morphology throughout plant development and across angiosperm evolution and plays a major role in determining floral organ shape.
Methods
Filament vasculature light microscopy
Costus and Musa filaments were fixed and dehydrated using Sorenson's solution and ethanol series, respectively. Plant material was embedded in paraffin and slides were prepared from 10 μm sections. Johansen's Safranin and Fast Green staining technique was used and light microscopy was performed using AxioImagerM1 373.
RNA extraction and Illumina Library preparation and processing
Musa basjoo (UCBG) and Costus spicatus (Specht Lab Greenhouse, UCB) flowers were collected and floral organs were carefully dissected. Immediately following dissection, floral organs were flash frozen in liquid nitrogen and stored at −80°C. Organ-specific total RNA extraction was performed using INVITROGEN Plant RNA Reagent, according to the manufacture's guidelines. Illumina libraries were prepared using the TruSeq RNA sample prep kit v2. Two libraries each were prepared from Musa basjoo filament and free petal RNA and Costus spicatus filament RNA and multiplexed using barcoding set A. Samples were run in a HiSeq2000 at IIGB HT Sequencing Facility at the University of California, Riverside.
Raw reads were trimmed to remove adapters and regions of poor quality with cutadapt. Costus spicatus sequences were assembled into a reference transcriptome using Trinity29.
Two independent replicates of RNA-seq data for filaments in Brassica rapa B3 were generated from greenhouse-grown plants, from which the central half of the filament was stored in RNAlater until RNA extraction was performed. FPKMs were generated according to ref. 30. All NGS libraries will be made publically available at SRA on NCBI.
Polarity gene orthologs in Arabidopsis thaliana, Musa acuminata and Costus spicatus
The polarity gene network for Arabidopsis thaliana is integrated elsewhere20. Sequences for each of the described Arabidopsis thaliana polarity genes were downloaded from NCBI: KANADI genes ATS, (NM_001125891.1) KAN2 (NM_102957.3), KAN3 (NM_117878.2), KAN4 (NM_121662.2); one AGONAUTE gene (AGO1, NM_001198240.1); YABBY genes INO (NM_102191.5), YAB5, (NM_179749.2), YAB2 (NM_001084021.1), AFO (NM_130082.3), CRC (NM_105585.2), YAB3 (NM_116235.2); two ASYMETRIC LEAVES, AS1(NM_129319.3) and AS2 (NM_105235.4); NUB (NM_101210.2), JAG (NM_105519.3); PHB (NM_129025.3), REV (NM_125462.3) and PHV (NM_102785.4).
Coding sequences (predicted CDS) for Musa acuminata genes were downloaded from the CIRAD website (Musa acuminata genome CIRAD Website - http://banana-genome.cirad.fr/download.php; D'Hont et al. 2012) and reciprocal BLAST was used to search for Musa acuminata orthologs of the Arabidopsis thaliana polarity genes listed above. Costus spicatus orthologs were identified using reciprocal BLAST against the assembled transcriptome using the Musa acuminata gene list.
In order to confirm BLAST results, a gene tree was built for the YABBY gene family. A multiple sequence alignment was performed using genes downloaded from NCBI, as well as those collected from the Musa acuminata genome. jModelTest was used and GTR was chosen as the best fit model. A maximum likelihood analysis was performed using PHYML (http://www.atgc-montpellier.fr/phyml/) with 100 bootstrap replicates.
Polarity gene expression
GSNAP was used to align Musa basjoo trimmed reads to annotated CDS from the published Musa acuminata genome and Costus spicatus trimmed reads to the Costus spicatus reference transcriptome. The expression of polarity genes was estimated using eXpress in units of FPKM (frequency per kilobase of exon per million aligned reads). Replicates were independently processed and gene expression was compared between libraries for consistency.
In order to confirm transcriptome gene expression of genes of interest, Real Time PCR (RT-PCR) was carried out. Total RNA was extracted from fresh tissues of labellum (fused petaloid staminodes) and the petaloid filament of Costus spicatus, as well as from the single free petal and the cylindrical filament of Musa acuminata (UC Davis) using INVITROGEN Plant Reagent. Total RNA was treated with Ambion TURBO DNA according to the rigorous treatment for abundant DNA contamination, following the protocol provided by the manufacturer. cDNA was generated from 1 μg of total RNA using SuperScript®, according to the provided protocol. Reactions without reverse transcriptase (no-RT) were performed as a control for genomic DNA contamination. Actin was used as a positive control for cDNA synthesis (Supplement Figure S2).
RT-PCR primers were designed using Primer3 (http://frodo.wi.mit.edu/) based on Musa basjoo and Costus spicatus transcripts. ACTIN1 was used as an internal control for the RT-PCR reaction. Expression was confirmed for three genes for both Musaacuminata (Ma) and Costus spicatus (Cs): YABBY2/5 orthologous genes; AS1 orthologs and a homolog of KAN2/3. Musa acuminata primers used were the following: MaACTIN1 forward TCCATCATGAAGTGCGATGT, MaACTIN1 reverse CTCTGCTTTTGCAATCCACA; MaYABBY2/5 forward AGCATTGTAGCAGTGCGATG, MaYABBY2/5 reverse GGACGCATAGGCAGCATAAT; MaAS1 forward AGCGGTGGAGATCTGAAGAG, MaAS1 reverse CGCGATCTTCTTCCACTTGT; MaKAN2/3 forward CTTCCCAACACAGCCAGATT, MaKAN2/3 reverse CGAAATTGGAGGTGGAAGAA. Costus spicatus primers, for orthologous genes, were the following: CsACTIN1 forward GCATGAGCAAGGAGATCACA, CsACTIN1 reverse CAAACATGACTTGGGTGTGC; CsYABBY2/5 forward CGGTTAGTGTGCCAGGAAAT, CsYABBY2/5 reverse CATGGGGAGCATCTGTTCTT; CsAS1 forward GATCCGAAGAGGATGCGATA, CsAS1 reverse ACTTGTTTCCGTGCTTTGCT; CsKAN2/3 forward TTAGGAGGCCATGAGAGAGC, CsKAN2/3 reverse GGATTAACCTCGGGCAAGTT. Primers were tested for single band amplification and primer dimer prior to the RT reaction (Supplement Figure S2).
RT-PCR was performed with iQ SYBR Green Supermix kit (BIO RAD) for a 20 μl reaction (0,75 μl of 10 mM primers forward and reverse; 10 μl 2× iQ™ SYBR® Green Supermix; 5 μl of a 1:5 cDNA solution; and 3,5 μl H2O). The cycling protocol was as follows: 1× initial denaturation time at 95°C for 2 minutes, followed by denaturing and annealing-extension cycle 95°C for 10 seconds and 55°C for 30 seconds, 44×; and a melting curve from 65°C–95°C in 0.5°C increments, for 10 seconds. A standard curve (1:1; 1:10; 1:100; 1:1,000; 1:10,000) was carried out for each primer pair. Delta-Ct values are show in relation to ACTIN1 levels for all studied genes.
RNA in situs hybridization of YABBY2/5
Developing inflorescences of Costus spicatus were collected and manually dissected to remove late developing flowers and flower bracts. Immediately after dissection, flowers were placed in fresh, RNAse-free 3.7% FAA and vacuum infiltrated for 10 minutes. After complete infiltration and fixation, inflorescences were embedded in paraffin and 12 um sections were made using a manual microtome. Sense and anti-sense probes were synthesized in vitro, based on a unique 150 bp fragment of YABBY2/5 cloned into a pJET1.2 vector. T7 promoters were used for in vitro transcription with Roche DIG RNA labeling kit according to provided recommendations. Probes were precipitated, ressuspended on formamide and checked for degradation using agarose gel and Nanodrop measures. Probes were stored at −80C until further use. The in situ hybridization was performed as described elsewhere32.
Environmental scan electron microscopy
Flowers of Costus spicatus (Costaceae) were collected and immediately dissected. Fragments of fresh C. spicatus stamen tissue were mounted on carbon tape. Images were taken from abaxial and adaxial surfaces of fresh stamen filament. Images were generated using a TM-1000 scan electron microscope at the Electron Microscopy Lab, University of California, Berkeley.
References
Goethe, J. W. The Metamorphosis of Plants. (MIT Press, London, 2009).
Goto, K., Kyozuka, J. & Bowman, J. L. Turning floral organs into leaves, leaves into floral organs. Cur Opin in Gen & Dev. 11, 449–456 (2001).
Pelaz, S., Tapia-López, R., Alvarez-Buylla, E. R. & Yanofsky, M. F. Conversion of leaves into petals in Arabidopsis. Cur Biol. 11, 182–184 (2001).
Litt, A. & Kramer, E. M. The ABC model and the diversification of floral organ identity. Sem in Cell & Dev Biol. 21, 129–137 (2010).
Lönnig, W. E. Goethe, sex and flower genes. Plant Cell. 6, 574–577 (1994).
Mc Connell, J. R. & Barton, K. M. Leaf polarity and meristem formation in Arabidopsis. Development. 125, 2935–2942 (1998).
Sawa, S., Ito, T., Shimura, Y. & Okada, K. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell. 11, 69–86 (1999a).
Pekker, I., Alvarez, J. P. & Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 17, 2899–2910 (2005).
Toriba, T. et al. Distinct regulation of adaxial–abaxial polarity in anther patterning in rice. Plant Cell. 22, 1452–1462 (2010).
Baker, S. C., Robinson-Beers, K., Villanueva, J. M., Gaiser, J. C. & Gasser, C. S. Interaction among genes regulating ovule development in Arabidopsis thaliana. Genetics. 145, 1109–1124 (1997).
Villanueva, J. M. et al. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes & Dev. 13, 3160–3169 (1999).
Léon-Kloosterziel, K. M., Keijzer, C. J. & Koornneef, M. A seed shape mutant of Arabidopsis that is affected by integument development. Plant Cell. 6, 385–392 (1994).
McAbee, J. M. et al. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. The Plant J. 46, 522–531 (2006).
Talbert, P., Adler, H.-T., Parks, D. W. & Comai, L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 121, 2723–2735 (1995).
Waites, R. & Hudson, A. phantastica: a gene required for dorsiventrality of leaves in Antirrhinum majus. Development. 121, 2143–2154 (1995).
Kidner, K. A. & Timmermans, M. C. P. Mixing and matching pathways in leaf polarity. Cur Opin in Plant Biol. 10, 13–20 (2007).
Juarez, M. T., Twigg, R. W. & Timmermans, M. C. Specification of adaxial cell fate in maize leaf development. Development. 131, 4533–4544 (2004).
Alvarez-Buylla, R. E. et al. [Flower Development]. The Arabidopsis Book. (The American Society of Plant Biologists, 2010). http://www.bioone.org/doi/full/10.1199/tab.0127 Date of access: 15/12/2013.
D'Hont, A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 488, 213–219 (2012).
Eshed, Y., Baum, S. F. & Bowman, J. L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 99, 199–209 (1999).
Sarojam, R. et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. The Plant Cell. 22, 2113–2130 (2010).
Yamada, T. et al. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. The Plant J. 67, 26–36 (2011).
Floyd, S. K., Zalewski, C. S. & Bowman, J. L. Evolution of class-III homeodomain-leucine zipper genes in streptophytes. Genetics. 173, 373–388 (2006).
True, J. R. & Carroll, S. B. Gene co-option in physiological and morphological evolution. Ann Rev Cell and Dev Biol. 18, 53–80 (2002).
Tomescu, A. Megaphylls, microphylls and the evolution of leaf development. Trends Plant Sci. 14, 5–12 (2008).
Harrison, C. J. et al. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 434, 509–514 (2005).
Nakayama, H., Yamaguchi, T. & Tsukaya, H. Expression patterns of AADL, A CRABS CLAW ortholog in Asparagus asparagoides (Asparagaceae), demonstrate a stepwise evolution of CRC/DL subfamily of YABBY genes. Am. J. Bot. 97, 591–600 (2010).
Nakayama, H., Yamaguchi, T. & Tsukaya, H. Acquisition and diversification of cladodes: leaf-like organs in the genus Asparagus. The Plant Cell. 24, 929–940 (2012).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biothec. 29, 644–652 (2011).
Schnable, J. C. & Freeling, M. qTeller: a pipeline for comparing gene expression using community-generated RNA-seq data. PLoS-One. Accepted for publication (2013). http://qteller.com/ Date of access: 22/03/2014.
Smith, J. F., Kress, W. J. & Zimmer, E. A. Phylogenetic analysis of the Zingiberales based on rbcL sequences. Ann. Miss. Bot. Garden. 80, 620–630 (1991).
Bartlett, M., Kirchoff, B. & Specht, C. D. Epi-illumination microscopy coupled to in situ hybridization and its utility in the study of evolution and development in non-model species. Development Genes and Evolution 218, 273–279 (2008).
Acknowledgements
The authors would like to thank E. Sandoval from the UC Davis Greenhouses for providing Musa acuminata material, H. Forbes from the UC Botanical Gardens for support on obtaining Musa basjoo material, X. Chen and the Chen lab (UC Riverside) especially Shaofang Li and Yu Yu for preparing and sequencing miRNA and RNA libraries, L. Feldman and K. Jiang for assistance with Real-Time PCR. S. Ruzin and D. Schichnes for assistance with microscopy and M. Bartlett for permission to use images of Costus flower bud. Research contributing to this manuscript was supported by NSF Doctoral Dissertation Improvement Grant (DEB 1110461; AMRA and CDS), NSF IOS 0845641 (CAREER to CDS), the Hellman Family Faculty Fund (CDS) and UC MEXUS-CONACYT 12-571 to ERAB and CDS. AMRA was supported by a CAPES/Fulbright Fellowship and ERAB was supported by the Miller Institute for Basic Research in Science, University of California, Berkeley, CONACyT (180098; 180380) and DGAPA, UNAM (IN204011-3; IN226510-3).
Author information
Authors and Affiliations
Contributions
A.M.R.A. designed and performed experiments, analyzed data and prepared the manuscript; R.Y. collected and analyzed data; J.C.S. analyzed bioinformatics data; E.A.-B., C.D.S. and M.F. gave conceptual advice and edited the manuscript. E.A.-B. contributed with the central hypothesis and experimental design.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
de Almeida, A., Yockteng, R., Schnable, J. et al. Co-option of the polarity gene network shapes filament morphology in angiosperms. Sci Rep 4, 6194 (2014). https://doi.org/10.1038/srep06194
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06194
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.