Abstract
ARGONAUTE1 (AGO1) encodes a key component of the complexes mediating microRNA (miRNA) function in Arabidopsis. To study the regulation, action and interactions of AGO1, we conducted a genetic screen to identify second-site mutations modifying the morphological phenotype of ago1-52, a partial loss-of-function allele of AGO1. Unlike null ago1 mutations, the hypomorphic ago1-52 allele does not cause lethality or sterility; however, ago1-52 does produce a morphological phenotype clearly distinct from wild type. In our screen for modifiers of ago1-52, we identified suppressor mutations that partially restore wild-type morphology in the ago1-52 background and we termed these mas (morphology ofargonaute1-52suppressed). We focused on 23 of these putative suppressors. Linkage analysis of the mas mutations together with sequencing of the AGO1 gene in genomic DNA and cDNA from ago1-52mas plants indicated that 22 of the mas lines contain extragenic suppressors and one contains an intragenic suppressor that affects splicing of ago1-52. In the presence of the wild-type allele of AGO1, most of the mas mutations cause a mild or no mutant phenotype on their own, indicating that the ago1-52 mutant may provide a sensitized background for examining the interactions of AGO1.
Similar content being viewed by others
Introduction
The existence of RNA molecules with repressor1 or activator2 functions was proposed in the first models of gene expression regulation. Despite these early predictions, however, experimental evidence of the existence of chromosomally encoded small regulatory RNAs was not published until the last two decades of the XX century in prokaryotes and eukaryotes, with the discovery of MicF3 and lin-44,5, respectively. lin-4 belongs to a class of small RNAs that has received more attention in recent years, the microRNAs (miRNAs), single-stranded molecules of about 22 nt in length that hybridize by complementarity to their mRNA targets and then induce the degradation and/or attenuation of the translation of these mRNA targets. These processes take place in the cytoplasm, in ribonucleoprotein complexes named RISC (RNA-Induced Silencing Complexes), whose catalytic component in all species studied so far is a protein of the ARGONAUTE (AGO) family6,7,8. Since the discovery of lin-4, hundreds of eukaryotic genes have been found to be negatively regulated by miRNAs in many organisms, including humans and thousands have been predicted as miRNA targets using many different algorithms.
A collection of Arabidopsis thaliana (hereafter, Arabidopsis) mutants with morphologically abnormal leaves was obtained in the laboratory of J.L. Micol9,10 and 41 of these mutants were dubbed incurvata (icu), because of their upwardly curved leaves11,12,13,14,15. We positionally cloned the icu3, icu8, icu9-1, icu9-2 and icu15 mutations16,17, which were found to be novel loss-of-function alleles of the genes encoding HASTY (HST)18, HYPONASTIC LEAVES1 (HYL1)19, ARGONAUTE1 (AGO1)20 and HUA ENHANCER1 (HEN1)21; these proteins are known components of the miRNA pathway22,23.
The above mentioned icu mutants were renamed as hst-21, hyl1-12, ago1-51, ago1-52 and hen1-13, respectively and intercrossed and crossed to dcl1-9 (dicer-like1-9)24, a line carrying a mutant allele of DCL1, another gene encoding a component of the miRNA machinery. All of the double mutant combinations obtained in this way showed a strong, synergistic phenotype16. This observation led us to presume that loss-of-function alleles of HST, HYL1, AGO1, HEN1 or DCL1 would provide a sensitized genetic background, useful for a second-site mutagenesis aimed to identify novel genes directly or indirectly related to the miRNA pathway.
To study the regulation, action and interactions of AGO1, we decided to perform a second-site mutagenesis with the ago1-52 mutant. The ago1-52 hypomorphic allele causes a morphological phenotype easily distinguishable from wild type16 and only partially reduces viability and fertility, unlike null ago1 alleles, which are completely sterile20. ago1-52 carries a G→A transition mutation 12 bp upstream of the 3′ end of its 21th intron17. The mutation creates a splicing acceptor signal that causes mis-splicing and gives rise to an mRNA 10 nt longer than that of the wild type; the ago1-52 mutant mRNA is translated into a mutant protein with 15 aa different than those of the wild type at the carboxyl terminus and with 55 fewer residues than the wild type (Figure S1). For the mutagen, we chose ethyl methanesulfonate (EMS) for its strong mutagenicity and its capacity to generate hypomorphic alleles, which might provide a broader range of suppressor mutations than null alleles25,26,27.
Results and Discussion
Morphological phenotype of the ago1-52 mutant
The ago1-52 recessive mutation causes a pleiotropic phenotype with complete penetrance and variable expressivity (Figure 1). At 21 days after stratification (das) the ago1-52 plants have an average of 4.07 ± 0.25 vegetative leaves, in contrast to the Ler plants, which have an average of 8.33 ± 0.84 leaves (n = 30). As in other ago1 mutants, leaves of ago1-52 show no clear boundary between petiole and lamina17. Juvenile ago1-52 leaves (the first three) exhibit abaxial trichomes, which are seen in the wild type only in adult leaves (fourth and following). Leaf adaxialisation is apparent in some ago1-52 plants, which exhibit different extents of radialisation of the two first leaves17. The ago1-52 plants do not exceed 20 cm in height (13.79 ± 1.82 cm in ago1-52 versus 25.5 ± 1.73 cm in Ler; n = 20) and possess a short, compact inflorescence. The ago1-52 plants have shorter siliques than the wild type (5.77 ± 1.52 mm in ago1-52versus 10.10 ± 1.54 mm in Ler; n = 30) and have fewer seeds than wild type (10.79 ± 8.49 in ago1-52versus 44.14 ± 16.92 in Ler; n = 30).
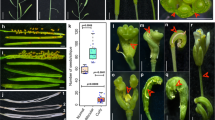
Some phenotypes of the ago1-52 mutant.
(a–c) Rosettes from (a) the wild type Ler and (b, c) ago1-52. An arrow in c highlights a trumpet-shaped, radialised leaf. (d) Detail of a radialised ago1-52 leaf. (e, f) Terminal region of the inflorescences of (e) Ler and (f) ago1-52. Scale bars: 1 mm. Pictures were taken (a–d) 21 das and (e, f) 42 das.
Screen design and mutagenesis
To isolate modifiers of the morphological phenotype of the ago1-52 mutant, we mutagenised homozygous ago1-52 plants with EMS. The procedure followed to isolate and characterise new mutants (Figure 2) was similar to that described by Berná et al.9. Approximately 67,500 seeds of ago1-52 were sent to Lehle Seeds (www.arabidopsis.com) to be mutagenized. Mutagenized seeds (M1 generation) were sown on soil and grown in a growth chamber to obtain M2 seeds, which were harvested in bulk to form 15 parental groups; these groups were then sent to our laboratory for screening.
Estimation of the efficiency of the mutagenesis
One of the methods used in Arabidopsis to estimate the efficiency of mutagenesis is based on determining the frequency of initial cells with mutations (P) and the mean number of mutations per initial cell (M)28. The values of P and M are calculated from the frequency of M1 plants that show siliques with some sort of mutation (ma) or from the frequency of siliques with mutations (mb), which, in turn, are determined according to the appearance of easily-observable mutant phenotypes in the silique, phenotypes such as embryo lethality or albinism, which occur very rarely in wild-type populations. P and M are then calculated as follows28,29:


where n is the mean number of initial cells, whose most widely accepted value is 2–3. About 50 M1 plants are usually examined and 5–6 siliques in each plant, to count the presence of albino embryos. Any mutagenesis with EMS that leads to a P value of above 0.3 can be considered effective. Indeed, using the equation in which the P value was defined, it can be seen that, assuming n = 2, P will = 0.3 when half the M1 plants show at least one mutation that causes embryo albinism (ma = 0.5).
Ninety siliques from 18 M1 plants were examined and albino embryos were found in all of them. As a consequence, the proportion of siliques revealing the existence of a mutation causing albinism was mb = 90/90 = 1, which implies P = 1. From the above, we can deduce that the frequency of M1 plants with siliques showing some mutation was ma = 1. In other words, all the M1 plants were carriers of at least one mutation causing embryo albinism. This value is surprisingly high, since in a similar mutagenesis carried out by Lehle Seeds for J.L. Micol on a wild type Ler genetic background, the value of P was 0.549,30. Perhaps in the sensitized ago1-52 genetic background many phenotypes, including albinism, arise that are much less visible in a wild-type background.
Of the M2 seeds studied, 38% showed no or abortive germination. The causal mutations for such lethal phenotypes in mature embryos (seeds) or seedlings seem to occur in all the M1 parental lines. This conclusion is based on the assumption that almost all these mutations were recessive, as is common for hypomorphic or null mutations and that each of them was homozygous in one of the three M2 seeds representing by each average M1 parental.
The number of albino seedlings also indicates the presence of mutations in the genomes of the plants screened. We found one case of albinism per every 91 M2 plants that did not show early lethality, which indicates that the mutagenesis was very effective. We use the term albinism here to refer to the absence of a green colour in the cotyledons of the seedlings, some of which were completely white and others yellowish.
Characterisation of the mutants
Phenotypic classification of putative double mutants
The screen for modifiers of the phenotype of ago1-52 was carried out in two steps, using eight and seven parental groups, respectively. We first screened 36,810 M2 seeds, the progeny of 10,264 M1 plants (parental groups P1–P8; Table 1) sowing seeds in plates in a controlled number with Ler and ago1-52 seeds as controls (see Methods). Whereas the ago1-52 and Ler seeds germinated at a rate of 93.0% and 97.2%, respectively and developed into viable plants, we observed no or abortive germination in 38.0% (14,121) of the M2 seeds sown. Such a high percentage of early lethality shows the sensitivity of the ago1-52 genetic background to EMS. Another indicator of the efficiency of the mutagenesis was the large number (249) of seedlings with albino cotyledons and no or only rudimentary leaves; all these seedlings died before 21 das (Table 1).
About 11% of the M2 seeds sown produced plants that survived more than 21 das and were considered double mutants because they showed unexpected phenotypes or an increased or reduced mutant phenotype compared to that of ago1-52 plants. Among the 4,189 double mutants isolated, 3,366 exhibited lethality after 21 das and died before completing their life cycle, 521 completed their life cycle but did not produce seeds and only 302 produced M3 seeds (Table 1).
M2 double mutants of the most represented class showed a strong synergistic phenotype, in some cases reminiscent of those of the double mutant combinations of mutations in two miRNA machinery genes and almost all showed a lethal phenotype. We also defined six phenotypic classes based on morphological traits, all of which included viable double mutants (Table 2 and Figure S2). Although some showed more than one of the traits considered characteristic of each class, we provisionally assigned each mutant to only one class. Plants showing leaf variegation or generalized chlorosis were assigned to the “De-pigmented plants” class. The “Severe Ago1-52 phenotype” and “Weak Ago1-52 phenotype” classes included plants in which the phenotype of ago1-52 was accentuated or alleviated, respectively. The “Reticulate leaves” class included plants with leaves in which the veins stood out because of their colour compared with a pale lamina. Plants with small vegetative leaves or short petioles were included in the “Compact rosette” class. The “Other phenotypes” class included a number of mutants with phenotypes that could not be categorized into the other classes.
Analysis of the transmission of the suppression of the phenotype of ago1-52
To study the inheritance of the suppressed phenotype, we examined the progeny of the isolated M2 plants. We sowed several tens of M3 seeds derived from each M2 plant self-pollination. Transmission of the mutant phenotype was unequivocally Mendelian in only 92 of the 302 M2 putative mutants that were fertile. We observed variable expressivity in 104 M3 families, a phenotype weaker than that of their M2 parentals in 21 M3 families and in 85 families the M2 phenotype did not reappear in the M3 generation.
Of the 92 M2 putative double mutants that showed complete penetrance and almost invariable expressivity in the M3 progeny, we focused on studying 17 lines of the 21 that we initially assigned to the class we denominate “Weak Ago1-52 phenotype” (Figure 3c-s). All these M3 lines exhibited suppression of the ago1-52 phenotypes during their vegetative and reproductive development. Leaves of the M3 plants were less spatulated than those of ago1-52, with a well-defined boundary between petiole and lamina. No radialized leaves were seen in these suppressor lines, which exhibited in addition more vegetative leaves than ago1-52. Suppression was also shown by these plants after bolting: they exhibited increased stature and number of stems compared to ago1-52, from which they also differ in having a less compact inflorescence (Figure 4). The number of seeds in the siliques of these suppressor lines was also higher than that of ago1-52 plants.
Examples of other phenotypes of the suppressor lines identified in this work.
Plant height of the (a) ago1-52 mas1-1 (P1 5.33), (b) ago1-52 mas2-1 (P2 11.1) and (c) ago1-52mas3-1 (P8 14.1) double mutants was intermediate between those of the wild type Ler and the ago1-52 single mutant. As shown in the insets, the structure of the terminal region of the inflorescence in the double mutants was also intermediate between those of Ler (Figure 1e) and ago1-52 (Figure 1f). Pictures were taken (a) 54 das, (b) 43 das and (c) 48 das. Scale bars: (a–c) 1 cm and (insets) 1 mm.
We hypothesized that partial suppression of the phenotype of the ago1-52 mutation shown by these lines may have been caused by loss of function in genes whose products have an antagonistic effect on AGO1, or by gain of function alleles of genes that act together with AGO1 in the silencing of miRNA targets. We did not study the lines of the remaining phenotypic classes, most of which were poorly viable and semi-fertile or sterile. We also did not study lines with phenotypes that seemed merely additive to that of ago1-52, since the corresponding double mutants were considered likely carriers of novel alleles of genes involved in processes unrelated to AGO131.
We also screened an additional set of about 20,000 M2 seeds later, belonging to the P9–P15 parental groups. In this case we sowed the seeds at high density (500 seeds for plate) in top agar and directly looked for plants with a weak Ago1-52 phenotype. We found six additional suppressor lines in this way (Figure 3t–y), which were studied together with the 17 lines already chosen in our first screen.
To exclude the possibility of contamination with wild-type seeds, we sequenced the AGO1 gene in all the suppressor lines, confirming the presence of the original, homozygous ago1-52 mutation in all of the lines when genomic DNA was used as template. We also confirmed the effect of ago1-52 on splicing when cDNA was used as template. The genomic and cDNA sequences revealed that one of the lines, P9 3.3, carried an intragenic suppressor mutation (which we termed ago1-52S), since it carries a G to A transition adjacent to the ago1-52 mutation (Figure 5a). Sequencing of AGO1 cDNA from P9 3.3 showed that the splicing acceptor (AG) site of the 21st intron had been shifted 1 nt downstream, causing the inclusion in the mutated mRNA of 9 nt that are intronic in the wild type, instead of 10 nt as occurs in ago1-52 mRNA; this restored the wild-type reading frame, although inserting three additional codons that were not present in the wild-type mRNA (Figure 5b, c).
The P9 3.3 line carries an intragenic suppressor mutation of ago1-52.
(a) Structure of the AGO1 gene with indication of the nature of the ago1-52 and ago1-52S mutations (green arrows and red letters). Positions of the start (ATG) and stop (TGA) codons are also indicated. Exons are shown as boxes and introns as lines between boxes. Open boxes represent untranslated exon sequences. (b) Effects of the ago1-52 and ago1-52S mutations on splicing and mRNA translation. Intron sequences are shown in small case and exon sequences in capital letters. The preferred splicing acceptor sites are boxed in green. Amino acids different from those of the wild type are shown in red. The asterisk indicates a premature stop codon. (c) Sequencing electropherograms obtained from Ler (AGO1), ago1-52 and P9 3.3 genomic DNA (gDNA; top) and cDNA (bottom). Nucleotide sequences that are intronic in the AGO1 wild type allele but exonic in the ago1-52 mutant and the ago1-52ago1-52S double mutant are boxed in orange.
We named the genes whose mutations suppressed the morphological phenotype of ago1-52 as MORPHOLOGY OF argonaute1-52 SUPPRESSED (MAS). We backcrossed the ago1-52mas suppressor lines to Ler twice. We transferred 1–5 F1 plants derived from each backcross into pots to complete their life cycle and collected the F2 seeds resulting from self-pollination. Between 150 and 450 seeds of several F2 families from each line were sown and the morphological phenotypes of the corresponding plants were examined to identify ago1-52/ago1-52;mas/mas and AGO1/AGO1;mas/mas plants.
We assumed that, as is usual for EMS-induced mutations, the suppressor mutations would be recessive, in most if not all cases. Hence, we expected to find four phenotypic classes in the F2 progeny: (1) the wild type (AGO1/-;MAS/-) class, (2) that of the mutation to be identified (AGO1/-;mas/mas), (3) Ago1-52 (ago1-52/ago1-52;MAS/-) and (4) the double mutant (ago1-52/ago1-52;mas/mas) class. As seen in Table S2, some of the mas mutations did not show any visible morphological phenotype in the AGO1/-;mas/mas genotype (P1 5.33, P2 11.1, P4 22.2, P5 11.1 and P8 14.1) whereas others do (P7 13.1, P7 23.1, P7 26.1 and P8 25.1).
We also note that in 6 of the 9 F2 derived from the first backcrosses to Ler we found mutant phenotypes additional to those initially seen in the M2 (Table S2). This suggests the presence of mutations in different genes in the M2 individual initially selected as a double mutant and reinforces the importance of backcrossing to Ler several times to reduce the number of mutations not relevant to the phenotype. In addition, the proportions of the phenotypes that we observed in the F2 did not fit any known Mendelian segregation. No or abortive seed germination contributed to this, as well as lethality and plants showing phenotypes that had not been seen in the M2 or M3.
In all the lines studied except two (P5 11.1 and P7 26.1), the double mutant class is more numerous than that of the single mutants. Given that the mutations under study normalise the body architecture of the ago1-52 mutant, it is reasonable to suppose that they also increase its fertility. As previously shown, the number of seeds per silique of the ago1-52 mutant is 25% that of the wild type, which explains why the ago1-52/ago1-52;mas/mas plants are more numerous than ago1-52/ago1-52;MAS/- plants. The absence of the AGO1/-;mas/mas class in some of the lines suggests that plants of this genotype are included in the phenotypically wild-type class. As a consequence, the populations of the first three lines in Table S2 should fit a 12:3:1 segregation (AGO1/-;-/- : ago1-52/ago1-52;MAS/- : ago1-52/ago1-52;mas/mas), but they do not because of the much better viability of the suppressed plants compared with the plants showing the full ago1-52 mutant phenotype.
We chose for subsequent study the P1 5.33, P2 11.1 and P8 14.1 lines for several reasons. Lines P1 5.33 and P2 11.1 were the first to be isolated in our screen and, therefore, the first ones to be studied. Line P8 14.1 was chosen because it showed a more pronounced suppression of the phenotype of ago1-52 during the reproductive phase. In the F2 of the backcrosses involving P1 5.33, P2 11.1 and P8 14.1 we only found three phenotypic classes: entirely wild type phenotype, Ago1-52 and double mutants, which indicates that the suppressor mutations did not cause a mutant phenotype on their own, either as homozygotes or heterozygotes in the presence of the wild type allele of AGO1. We did not perform complementation analyses by intercrossing the suppressor lines. Rather, we first used linkage analysis to determine their genetic map positions (see examples in Table S3) and then crossed for allelism tests only the lines carrying mutations with neighbouring map positions. Linkage analyses demonstrated in all cases that the suppressor mutations are extragenic suppressors that map to chromosomes 2, 3, 4 or 5, or on chromosomes 1 but far from AGO1; the only exception was the above-mentioned ago1-52S intragenic suppressor mutation. The genetic and molecular characterization of each of these suppressors will be described elsewhere.
Concluding remarks
EMS has been widely used to induce mutations in forward and second-site genetic screens with many experimental organisms, but also to increase crop diversity. Further, EMS has been used in saturation mutagenesis in several TILLING (Targeting Induced Local Lesions in Genomes)32,33 projects with different animal and plant species, including Arabidopsis, because it causes randomly distributed point mutations and only rarely produces DNA rearrangements that could result in lethality26. Mutagenesis in Arabidopsis predominantly relies on EMS and its non-lethal dose treatment has been well established. Therefore, it was striking that our ago1-52 mutagenesis led to a very high percentage of lethality. EMS mainly induces alkylation of G residues, which then pair with T instead of with C; if these G:T mismatches are not repaired, they produce G/C to A/T transitions.
Universal repair mechanisms, such as the mismatch repair pathway that recognizes G:T mismatches, as well as repair of alkylated nucleotides by DNA glycosylases, act in Arabidopsis34,35,36. The apparently high sensitivity to EMS that we observed in the ago1-52 background could be the result of defects in these DNA repair mechanisms. If this were true, ago1-52 plants should accumulate mutations at a higher density than wild-type plants or other non-sensitive backgrounds under similar non-lethal EMS doses. Another interpretation of our results could be that since AGO1 affects many pivotal processes, these processes are all de-regulated in the ago1-52 mutant and second-site mutations in genes involved in other pivotal processes could result in lethality. Comparative analysis of whole-genome sequences of M2 plants obtained from either ago1-52 or wild-type plants both treated with EMS could settle this question.
Second-site mutagenesis screens are common strategies for finding functionally related genes. The pleiotropic phenotype of the ago1 mutants results from the alteration of many biological processes, which involve many genes that are directly or indirectly regulated by the miRNA pathway. It may, therefore, seem surprising that we obtained ago1-52mas double mutants with morphological phenotypes very close to that of the wild type. Suppressor mutations usually fall into two classes: informational suppressors and functional suppressors. Informational suppressors act through a generic mechanism, such as the transcription machinery, RNA processing and translation and they can also suppress a premature stop codon, modify aberrant splicing of mRNA or control mRNA translation or protein degradation. For example, in Caenorhabditis elegans, three kinds of informational suppression have been described so far: nonsense suppression, suppression by modified splicing and suppression by loss of nonsense-mediated decay. By contrast, functional suppressors act through mechanisms directly related to the process of interest37,38. Since informational suppressors generally are allele-specific but not gene-specific, we will test allele- and gene-specificity in all our mas mutations.
Methods
Plant material and growth conditions
Arabidopsis thaliana (L) Heynh. Landsberg erecta (Ler) and Columbia-0 (Col-0) wild-type accessions were obtained from the Nottingham Arabidopsis Stock Center (NASC; Nottingham, UK) and then propagated at our laboratory for further analysis. Seed sterilization and sowing, plant culture and crosses were performed as previously described9,39. In brief, seeds were sown on plates containing MS agar medium (half-strength Murashige and Skoog salts, 0.7% plant agar [Duchefa], pH 5.7 and 1% sucrose) and stratified (4°C in the dark) for 48 h and then transferred to either Conviron TC16 or TC30 growth chambers set to our standard conditions (continuous light at approximately 75 μmol·m−2·s−1, 20 ± 1°C, 60–70% relative humidity). When required, plants were transferred into pots containing a 2:2:1 mixture of perlite:vermiculite:sphagnum moss and grown in walk-in growth chambers set to our standard conditions.
Mutagenesis and mutant isolation
≈67,500 ago1-52 seeds (1 g) were sent to a commercial supplier, Lehle Seeds (http://www.arabidopsis.com), where they were mutagenised by immersion in a solution of 0.23% (v/v) EMS for 12 hours at 25°C, stratified for 7 days and then sown in pots. The M2 progeny obtained from selfed M1 plants was sent to us as 15 envelopes, each of which contained the pooled M2 seed progeny of a parental group: 1,283 M1 plants developed from seeds exposed to EMS.
Seeds were sown in 15 cm-diameter Petri dishes containing solid MS medium. Each dish was sown with 94 M2 seeds, together with 8 Ler and 8 ago1-52 seeds, which served as controls. Selection of putative double mutants was carried out 21 days after stratification (das) by eliminating all the M2 plants not showing clear morphological differences from ago1-52. All plants distinguishable from ago1-52 were presumed to be double mutants and they were transplanted into pots and allowed to complete their life cycles. Putative double mutants were given protocol numbers, as PN X.Y: PN indicates the corresponding parental group, X refers to the number of the plate where the mutant was isolated and Y is an ordinal assigned to each of the mutants found in a given plate.
Linkage analysis, RNA isolation and genomic DNA and cDNA sequencing
Low-resolution mapping of the suppressor mutations was performed by linkage analysis as described in Ponce et al.40,41. Genomic DNA was isolated as previously described42. Total RNA from 20–30 mg of rosette leaves, collected 21 das, was isolated using TRI Reagent (Sigma) and first-strand cDNA synthesis was performed as described in Jover-Gil et al.17. Genomic DNA and cDNA PCR amplification and sequencing were performed as previously described14,42. Primers for PCR amplifications and sequencing were ago1-52-27-F (5′-TTACCACGTTCTTTGGGATGAG-3′) and ago1-52-27-R (5′-GCAGTAGAACATGACACGCTTC-3′). The chromatograms shown in Figure 5 were obtained with Chromas Lite 2.1.1. (http://technelysium.com.au/).
References
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Britten, R. J. & Davidson, E. H. Gene regulation for higher cells: a theory. Science 165, 349–357 (1969).
Esterling, L. & Delihas, N. The regulatory RNA gene micF is present in several species of gram-negative bacteria and is phylogenetically conserved. Mol. Microbiol. 12, 639–646 (1994).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Höck, J. & Meister, G. The Argonaute protein family. Genome Biol. 9, 210 (2008).
Meister, G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459 (2013).
Peters, L. & Meister, G. Argonaute proteins: mediators of RNA silencing. Mol. Cell 26, 611–623 (2007).
Berná, G., Robles, P. & Micol, J. L. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742 (1999).
Serrano-Cartagena, J., Robles, P., Ponce, M. R. & Micol, J. L. Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261, 725–739 (1999).
Serrano-Cartagena, J. et al. Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156, 1363–1377 (2000).
Ochando, I. et al. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol. 141, 607–619 (2006).
Pérez-Pérez, J. M. et al. A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 51, 1661–1673 (2010).
Barrero, J. M., González-Bayón, R., del Pozo, J. C., Ponce, M. R. & Micol, J. L. INCURVATA2 encodes the catalytic subunit of DNA polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19, 2822–2838 (2007).
Esteve-Bruna, D., Pérez-Pérez, J. M., Ponce, M. R. & Micol, J. L. incurvata13, a novel allele of AUXIN RESISTANT6, reveals a specific role for auxin and the SCF complex in Arabidopsis embryogenesis, vascular specification and leaf flatness. Plant Physiol. 161, 1303–1320 (2013).
Jover-Gil, S., Candela, H. & Ponce, M. R. Plant microRNAs and development. Int. J. Dev. Biol. 49, 733–744 (2005).
Jover-Gil, S. et al. The microRNA pathway genes AGO1, HEN1 and HYL1 participate in leaf proximal-distal, venation and stomatal patterning in Arabidopsis. Plant Cell Physiol. 53, 1322–1333 (2012).
Telfer, A. & Poethig, R. S. HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125, 1889–1898 (1998).
Lu, C. & Fedoroff, N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin and cytokinin. Plant Cell 12, 2351–2366 (2000).
Bohmert, K. et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180 (1998).
Chen, X., Liu, J., Cheng, Y. & Jia, D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094 (2002).
Park, W., Li, J., Song, R., Messing, J. & Chen, X. CARPEL FACTORY, a Dicer homolog and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Current Biol. 12, 1484–1495 (2002).
Han, M. H., Goud, S., Song, L. & Fedoroff, N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101, 1093–1098 (2004).
Jacobsen, S. E., Running, M. P. & Meyerowitz, E. M. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126, 5231–5243 (1999).
Weigel, D. & Glazebrook, J. EMS mutagenesis of Arabidopsis seed. Cold Spring Harb. Protoc. 2006 (2006).
Kim, Y., Schumaker, K. S. & Zhu, J. K. EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 323, 101–103 (2006).
Qu, L. J. & Qin, G. Generation and identification of Arabidopsis EMS mutants. Methods Mol. Biol. 1062, 225–239 (2014).
Mednik, I. G. On methods evaluating the frequencies of induced mutations in Arabidopsis based on embryo-test data. Arabidopsis Inf. Serv. 26, 67–72 (1988).
Ivanov, V. I. Estimation of induced mutation rate in Arabidopsis. Arabidopsis Inf. Serv. 9, 31–32 (1973).
Quesada, V., Ponce, M. R. & Micol, J. L. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436 (2000).
Pérez-Pérez, J. M., Candela, H. & Micol, J. L. Understanding synergy in genetic interactions. Trends Genet. 25, 368–376 (2009).
McCallum, C. M., Comai, L., Greene, E. A. & Henikoff, S. Targeted screening for induced mutations. Nat. Biotechnol. 18, 455–457 (2000).
McCallum, C. M., Comai, L., Greene, E. A. & Henikoff, S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442 (2000).
Culligan, K. M. & Hays, J. B. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6 and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell 12, 991–1002 (2000).
Dany, A. L. & Tissier, A. A functional OGG1 homologue from Arabidopsis thaliana. Mol. Genet. Genom. 265, 293–301 (2001).
Garcia-Ortiz, M. V., Ariza, R. R. & Roldan-Arjona, T. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol. Biol. 47, 795–804 (2001).
Hodgkin, J. in WormBook (2005), pp. 1–13.
Fay, D. & Johnson, W. in WormBook (2006), pp. 1–4.
Ponce, M. R., Quesada, V. & Micol, J. L. Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 14, 497–501 (1998).
Ponce, M. R., Robles, P. & Micol, J. L. High-throughput genetic mapping in Arabidopsis thaliana. Mol. Gen. Genet. 261, 408–415 (1999).
Ponce, M. R., Robles, P., Lozano, F. M., Brotons, M. A. & Micol, J. L. Low-resolution mapping of untagged mutations. Methods Mol. Biol. 323, 105–113 (2006).
Pérez-Pérez, J. M., Ponce, M. R. & Micol, J. L. The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 134, 101–117 (2004).
Acknowledgements
The authors wish to thank J.L. Micol for helpful discussions and comments on this manuscript and for the use of his facilities. We wish also thank J.M. Serrano, F.M. Lozano, T. Trujillo and L. Serna for technical assistance. This work was supported by research grants from the Ministerio de Economía y Competitividad of Spain (BIO2008-01900) and the Generalitat Valenciana (Prometeo/2009/112) to M.R.P.
Author information
Authors and Affiliations
Contributions
M.R.P. conceived and designed the research. R.M.-P. and V.A. performed the research. M.R.P. and R.M.-P. wrote the article.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Micol-Ponce, R., Aguilera, V. & Ponce, M. A genetic screen for suppressors of a hypomorphic allele of Arabidopsis ARGONAUTE1. Sci Rep 4, 5533 (2014). https://doi.org/10.1038/srep05533
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05533
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.




 symbols indicate growth and selfing, respectively. The
symbols indicate growth and selfing, respectively. The  symbol indicates outcrossing or backcrossing.
symbol indicates outcrossing or backcrossing.

