Abstract
The basic leucine zipper transcription factor CCAAT/enhancer binding protein alpha (CEBPA) codes for a critical regulator during neutrophil differentiation. Aberrant expression or function of this protein contributes to the development of acute myeloid leukemia (AML). In this study, we identified two novel unrelated CEBPA target genes, the glycolytic enzyme hexokinase 3 (HK3) and the krüppel-like factor 5 (KLF5) transcription factor, by comparing gene profiles in two cohorts of CEBPA wild-type and mutant AML patients. In addition, we found CEBPA-dependent activation of HK3 and KLF5 transcription during all-trans retinoic acid (ATRA) mediated neutrophil differentiation of acute promyelocytic leukemia (APL) cells. Moreover, we observed direct regulation of HK3 by CEBPA, whereas our data suggest an indirect regulation of KLF5 by this transcription factor. Altogether, our data provide an explanation for low HK3 and KLF5 expression in particular AML subtype and establish these genes as novel CEBPA targets during neutrophil differentiation.
Similar content being viewed by others
Introduction
The CCAAT/enhancer binding protein alpha (CEBPA) is a master regulator of myeloid differentiation that drives hematopoiesis towards the granulocytic lineage1. The relevance of CEBPA in the maintenance of myeloid lineage is underlined by the fact that Cebpa-null mice lack the production of neutrophils and eosinophils2. Not surprisingly, CEBPA functions are frequently disrupted in particular subtypes of acute myeloid leukemias (AML), thereby contributing to the differentiation arrest seen in AML blast cells. Several groups reported aberrant CEBPA function due to intragenetic mutations in 5–10% of AML patients (10–19% in normal karyotype-AML) (Reviewed in3,4,5,6). Mutations in the CEBPA gene predominantly cause either a frame-shift at the N-terminus resulting in the truncated CEBPA isoform p30 with dominant negative function, or disrupt the basic zipper region at the C-terminus causing weakened DNA-binding (Reviewed in7,8). Furthermore, most AML patients harbor CEBPA mutations on both alleles and these bi-allelic CEBPA mutated patients have a favorable clinical outcome as compared to CEBPA-wild-type or patients with a mono-allelic mutation (Reviewed in5,6). Lastly, transcriptional repression mediated by leukemic fusion proteins or epigenetic alterations such as aberrant promoter hypermethylation further leads to low CEBPA expression in some AML patients9.
Our group recently reported a functional role for hexokinase 3 (HK3) and Krüppel-like factor 5 (KLF5) in the pathogenesis of AML10,11. HK3 (ATP:D-hexose 6-phosphotransferase) is a glycolytic enzyme most frequently expressed in myeloid cells and represents the dominant hexokinase in granulocytes accounting for 70–80% of total hexokinase activity10,12. We identified HK3 as a direct target of the transcription factors PU.1 and oncogenic fusion protein PML-RARA in acute promyelocytic leukemia (APL) characterized by the t(15;17) translocation. Inhibition of HK3 impairs neutrophil differentiation of APL cells and promotes cell death upon anthracycline treatment. We found significantly reduced HK3 expression not only in APL but also in other AML subtypes suggesting that additional mechanisms responsible for low HK3 levels in AML are operative10.
We and others found that members of the Krüppel-like factor (KLF) family are often deregulated in primary AML patient samples11. The KLF transcription factor family is composed of 17 members involved in diverse functions such as proliferation, self-renewal, differentiation and apoptosis13. Among the different KLFs inhibited in AML, KLF5 was found to be essential for granulocytic differentiation11,14. Low KLF5 expression in AML can partially be explained by hypermethylation of its promoter as shown in primary AML and in several AML cell line models14. Still unclear is how KLF5 is transcriptionally regulated during granulocytic differentiation.
In this study we describe two novel CEBPA target genes, the glycolytic enzyme HK3 and the transcription factor KLF5, both of which are significantly downregulated in CEBPA-mutated AML patients.
Results
Low HK3 and KLF5 expression is associated with CEBPA-mutated AML
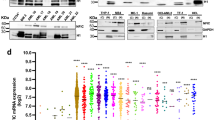
CEBPA is a master regulator of myeloid differentiation, whose expression peaks in the granulocyte-monocyte progenitor cell stage and diminishes during terminal granulocyte differentiation3. Most importantly, in different AML subtypes CEBPA expression or function is inhibited by a variety of mechanisms3,5,6. In an attempt to test whether HK3 and KLF5, two genes with a recently discovered role in APL differentiation10,11,14, are novel CEBPA target genes, we compared their expression in a total of 90 CEBPA-mutated or CEBPA wild-type AML patients. HK3 and KLF5 were below the detection limit in 11 and 16 AML patient samples, respectively. We compared HK3 mRNA expression in CD34+ samples (n = 4) to 74 CEBPA-wt and 5 CEBPA-mutated AML samples. KLF5 expression in 4 CD34+ samples was compared to 70 CEBPA-wt and 4 CEBPA-mutated AML samples. A detailed analysis of HK3 and KLF5 mRNA levels in primary AML (FAB M0-M7) patient samples revealed a significantly lower expression of HK3 and KLF5 in CEBPA-mutated as compared to CEBPA wild-type AML samples (p < 0.01) (Figure 1a–b). As seen for other CEBPA target genes identified by screening AML patient cohorts, our results suggest that disrupting CEBPA function impairs HK3 and KLF5 expression15. Next, we confirmed that HK3 and KLF5 expression is significantly lower in CEBPA-mutated, but not CEBPA wild-type patients from analysis of a second AML patient cohort (n = 154). Since these cohorts contained patients with CEBPA single (SM) or double mutations (DM), we could address whether single or double mutations alter HK3 or KLF5 expression16. We found that HK3 or KLF5 expression was not significantly different from patient's samples with either CEBPA SM or DM (Figure 1c–d). Taken together, our findings indicate that loss of HK3 or KLF5 expression correlated with CEBPA mutations, irrespective of single or double CEBPA mutations, whereas enhanced expression of HK3 or KLF5 correlated with wild-type CEBPA expression.
HK3 and KLF5 expression is significantly downregulated in CEBPA-mutated AML patients.
HK3 (a) and KLF5 (b) mRNA levels were measured by qPCR in total RNA extracted from primary AML (FAB M0-M7) blasts, CD34+ samples or granulocytes from healthy donors. Patient characteristics are summarized in Supplementary Table 1. HK3 (c) and KLF5 (d) levels in 154 patients from the Taskesen cohort with normal karyotype, expressing wild type CEBPA (WT), one allele mutated (SM) or two alleles mutated (DM). MWU: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
CEBPA-dependent induction of HK3 and KLF5 during neutrophil differentiation of APL cells
To experimentally test whether loss of CEBPA mRNA alters HK3 and KLF5 expression, we knocked down CEBPA in the two APL cell line models NB4 and HT93. This was accomplished using lentiviral vectors expressing two independent short hairpin (sh)RNAs targeting the CEBPA gene. Knockdown efficiency was evaluated by qPCR and Western blotting (Figures 2a, e and f). After 4 days of treatment with 1 μM ATRA to promote neutrophil differentiation, we observed significantly reduced HK3 and KLF5 mRNA levels in the NB4 and HT93 CEBPA knockdown cell lines as compared to the control cells (Figures 2b and c). We evaluated G-CSFR expression in CEBPA knockdown cells (Figure 2d) to validate inhibition of ATRA-induced neutrophil differentiation, while induction of CEBPE, a direct CEBPA regulated gene17, was measured as an additional control (Supplementary Figures S1a and b). Our results clearly demonstrate that the upregulation of HK3 and KLF5 during neutrophil differentiation of APL cells is CEBPA-dependent.
Genetic inhibition of CEBPA impairs HK3 and KLF5 upregulation during neutrophil differentiation of APL cells.
(a) NB4 or HT93 APL cell lines were stably transduced with pLKO.1 lentiviral vectors expressing non-targeting or two independent CEBPA-targeting shRNAs. APL control and CEBPA knockdown cells were differentiated with 1 μM ATRA for 4 days. Knockdown efficiency in NB4 (top panels) or HT93 (bottom panels) APL cells was validated by qPCR. CEBPA mRNA expression was normalized to the housekeeping gene HMBS and is shown as n-fold changes compared to untreated SHC002 control cells. HK3 (b) and KLF5 (c) mRNA expression in NB4 or HT93 APL cells was determined by qPCR and analyzed as in 2a. Impairment in granulocytic differentiation of CEBPA knockdown cells was shown by a reduction of the neutrophil marker granulocyte colony-stimulating factor receptor (GCSFR or CSF3R) (d) Data represent the mean ± s.d. of at least three independent experiments. (e), (f) CEBPA Knockdown efficiency at the protein level in NB4 and HT93 APL cells was confirmed by western blotting. GAPDH is shown as a loading control. MWU: *p < 0.05, **p < 0.01 and ***p < 0.001.
CEBPA binding to the HK3 or KLF5 promoters activates transcription
Our findings prompted us to investigate if HK3 and KLF5 are direct transcriptional targets of CEBPA. To this end, we first analyzed the genomic regions surrounding exon 1 of these genes for putative CEBPA binding sites using Matinspector 8.0. We identified two putative CEBPA binding sites in both the HK3 and the KLF5 genomic regions analyzed (Figure 3a and c). Chromatin immunoprecipitation (ChIP) revealed binding of CEBPA at positions +821/+2963 and −385/−1576 relative to the transcriptional start site on the HK3 and KLF5 genomic regions, respectively (Figures 3b and d). We next asked whether CEBPA activates HK3 and KLF5 transcription from these sites using promoter reporter assays. CEBPA co-expression with the different luciferase promoter reporters resulted in a significant, dose-dependent activation of HK3 promoter A (up to 37-fold) and KLF5 (up to 11-fold) transcription, respectively (Figures 3e and g). The HK3 promoter B containing the CEBPA binding site +2963 was only induced 1.5-fold (Supplementary Figure S2a). To confirm the specific CEBPA-induction of the promoter activity we mutated the CEBPA binding sites on the HK3 promoter A and the KLF5 promoter. Indeed, inactivating the CEBPA binding site in the HK3 promoter significantly inhibited transcriptional inhibition (Figure 3f). Surprisingly, however, introducing single or double CEBPA binding sites mutations in the KLF5 promoter did not attenuate transcriptional activity of the reporter construct (Supplementary Figure S2b).
CEBPA binds to and activates the HK3 and the KLF5 promoters.
Schematic representation of a 6 kb human HK3 (a) and a 5 kb human KLF5 (c) genomic regions retrieved from an online database. MatInspector software predicted two putative CEBPA binding sites (squares) in the DNA sequences analyzed. In vivo binding of CEBPA to these CEBPA consensus sites in the HK3 (b) or KLF5 (d) genomic regions was shown by chromatin immunoprecipitation (ChIP) in NB4 APL cells. As a negative control for the different pull downs, absence of GAPDH amplification is shown. *unspecific band, primer dimer. Two HK3 (e–f) and one KLF5 genomic region (g) containing the CEBPA binding sites were PCR amplified from genomic DNA of NB4 cells using proof reading Pfu DNA polymerase and cloned into the pGL4.10-basic vector. H1299 cells were transiently transfected with 40 ng of either HK3 promoter reporter construct A (e), construct A with mutated CEBPA binding site (f, wild-type GAAAGAC, mutated GGTCGAC) or the KLF5 promoter reporter construct (g), together with pcDNA3.1 empty vector or increasing concentrations (40–80–120 ng) (e,g) or 80 ng of CEBPA expression vector (f). The promoter activity is shown as relative light units (RLU) relative to pcDNA3.1 control transfected cells. Results are the means ± s.d. of at least triplicate transfections. MWU: **p < 0.01, ***p < 0.001.
Ectopic expression of CEBPA significantly induces HK3 and KLF5 transcription
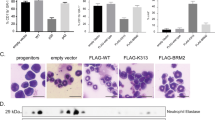
To confirm CEBPA-dependent transcriptional regulation of HK3 and KLF5, we overexpressed CEBPA in HT93 APL cells by transiently transfecting a CEBPA expression plasmid. Ectopic expression of CEBPA in HT93 cells resulted in a significant 5.5- and 12.3-fold induction of HK3 and KLF5 gene expression, respectively (Figures 4a and b). CEBPA transfection efficiency was assessed by qPCR and Western blotting (Figure 4d and e). In addition, induction of the direct CEBPA target CEBPE was measured as a positive control of CEBPA activity (Figure 4c). Next, we used K562 leukemic cell lines expressing 4-OHT inducible full-length CEBPA (CEBPA-p42-estrogen receptor (ER)), a truncated isoform (CEBPA-p30-ER), or a p42 isoform with a mutation preventing interaction with E2F and neutrophil differentiation (CEBPA-BRM2-ER). After 24 h of 4-OHT treatment, we observed a significant 2.5-fold transcriptional activation of KLF5 (p < 0.05) upon CEBPA-p42-ER activation only, but not in ER control, nor in CEBPA-p30-ER, or mutated CEBPA-BRM2-ER activated K562 cells (Figure 4f), suggesting that induction of KLF5 is a specific function of the full length CEBPA p42 isoform. Interestingly, HK3 mRNA was not detectable in K562 cells indicating that different HK isoforms may be active in these cells. Induction of CEBPE was measured as a positive control (Figure 4g). Altogether, our findings demonstrate that CEBPA is a crucial positive regulator of HK3 and KLF5 during granulocytic differentiation.
Ectopic expression of CEBPA activates HK3 and KLF5 transcription.
(a–e) HT93 cells were transiently transfected with pcDNA3.1 empty control or a CEBPA expression vector. HK3 (a) and KLF5 (b) mRNA expression was quantified by qPCR. Data were normalized to HMBS and are shown as n-fold regulation as compared to control transfected cells. Induction of CEBPE mRNA, a direct target gene of CEBPA, was measured as a positive control for CEBPA activity (c). Results are the means ± s.d. of at least triplicate transfections. CEBPA transfection efficiency was measured by qPCR (d) and western blotting (e). GAPDH is shown as a loading control. (f–g) Different CEBPA-ER fusion constructs were induced by treating the respective K562 cell lines with 5 μM Tamoxifen for 24 h. HK3 (f) or KLF5 (g) mRNA expression was quantified by qPCR as in 2a. Expression of the CEBPA target CEBPE was measured as positive control for CEBPA activation in wildtype CEBPA p42 expressing K562 cells. MWU: *p < 0.05, **p < 0.01.
Discussion
Reduced CEBPA expression or loss of function mutations are commonly observed in AML suggesting that the deregulation of CEBPA function is a major event in the development of AML18,19,20. Identifying new CEBPA target genes and associated CEBPA-dependent pathways in myeloid differentiation may provide novel insights into the differentiation block as well as into ancillary cellular functions found in AML. Interestingly, CEBPA has been described as a pivotal regulator of various metabolic processes in different cell types21. Nevertheless, whether CEBPA regulates metabolic activity in myeloid cells is of yet unclear. Our findings now link CEBPA to glycolysis via direct positive regulation of the glycolytic enzyme HK3. We have previously shown that HK3 expression is repressed in AML, particularly in APL, where low HK3 levels are the result of PML-RARA repression and low expression of its positive regulator PU.110. Our results demonstrate that CEBPA is an additional critical regulator of HK3 expression in AML and during neutrophil differentiation of APL cells. Furthermore, our findings show that the lowest HK3 expression is found in CEBPA mutated patients and may contribute to the differentiation block in this AML subtype.
In general, AML is defined by a block in differentiation and an increased proliferation of immature myeloid progenitors18. Additionally, loss of CEBPA-mediated cell-cycle arrest is crucial for the development of AML15,22,23. Since KLF5 regulates genes involved in cell cycle regulation and apoptosis, it may represent a relevant downstream effector of CEBPA-induced cell cycle arrest (reviewed in24). In the myeloid lineage, KLF5 induces differentiation and functions as a tumor suppressor and its low expression in AML is partially explained by epigenetic silencing11,14,25. Importantly, low KLF5 expression is associated with poor overall survival26. In this study, we suggest an additional mechanism for low KLF5 levels in AML that is loss of positive regulation of KLF5 due to impaired CEBPA function. Since mutating the CEBPA binding sites in the KLF5 promoter constructs did not change the responsiveness of these reporters to CEBPA expression, CEBPA may indirectly activate KLF5 transcription. The KLF5 promoter lacks a TATA box, but contains a GC-rich region that is activated by the transcription factor Sp127. Moreover, CEBPA can functionally interact with Sp1, e.g. in regulating the CD11c integrin gene28. We propose, that CEBPA induces KLF5 transcription, similarly to CD11c regulation, via Sp1.
In conclusion, we identified HK3 and KLF5 as novel CEBPA-regulated genes in AML and during APL differentiation underlining their tumor suppressor function in AML as well as their role in granulopoiesis.
Methods
Patient samples, cell lines and cell culture conditions
Fresh leukemic blast cells from untreated AML patients at diagnosis obtained at the Inselspital Bern (Switzerland) were classified according to the French-American-British (FAB) classification and cytogenetic analysis. All leukemic samples displayed a blast count of >90% after separation of mononuclear cells using a Ficoll gradient (Lymphoprep™, Axon Lab AG, Switzerland) as described previously10,11. PBMC and granulocytes obtained from normal, healthy donors were isolated using a Ficoll gradient.
The human acute promyelocytic leukemia (APL) cell lines NB4 and HT93 were maintained in RPMI-1640 (Sigma-Aldrich) with 10% fetal calf serum (FCS, Biochrom, AG), 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma-Aldrich) in a 5% CO2-95% air humidified atmosphere at 37°C.
For differentiation experiments, NB4 cells were seeded at a density of 0.2 × 106/ml and treated with 1 μM all-trans retinoic acid (ATRA, dissolved in DMSO, Sigma Aldrich) as indicated. Successful neutrophil differentiation was assessed by light microscopy using May-Grünwald-Giemsa (Merck, Darmstadt, Germany) stained cells and by surface expression of the differentiation marker CD11b (Immunotools). Briefly, 5 × 105 cells were collected, washed and incubated with monoclonal mouse phycoerythrin (PE)-labeled anti-human-CD11b for 20 min at 4°C. Fluorescence intensity was then measured by a FACS Calibur flow cytometer (Becton Dickinson, Basel, Switzerland) and analyzed using FlowJo Software.
K562-CEBPA-p42-ER, K562-CEBPA-p30-ER, K562-CEBPA-BRM2-ER and K562-ER were kindly provided by Prof. G. Behre and were maintained in RPMI-1640 supplied with 10% fetal calf serum (FCS), 50 U/mL penicillin and 50 μg/mL streptomycin in a 5% CO2-95% air humidified atmosphere at 37°C. K562 CEBPA-ER cells were differentiated as described15 by addition of 5 μM 4-OHT (Sigma-Aldrich).
Chromatin immunoprecipitation (ChIP)
ChIP was performed using the ChIP-IT Express Chromatin Immunoprecipitation Kit (ChIP-IT Express, Active Motif, Rixensart, Belgium) according to the manufacturer's recommendations. For immunoprecipitation, an anti-CEBPA antibody (sc-61X Santa Cruz, CA, US) was used. Antibodies against acetyl-histone H3 (Stratagene, La Jolla, CA, US) and IgG (PP64B, Upstate, Millipore) served as positive and negative controls, respectively. PCR was performed using the following primers: HK3 promoter A, F: GGGCTACTTGGGGATTGCTTCACA, R: ACGTGTCTTTCTCAAGGCCCAGC; HK3 promoter B, F: TCAGCCTTTCCAGTGCTCTCTCA, R: CTGTAGCAGTCAACCAGAAGGCCC; KLF5 promoter A, F: 5′-ACAGCATGGGGCTGCTTGGT -3′, R: 5′-TGCCTTCAGAGAATGGCTGATGAC -3′; KLF5 promoter B, F: 5′-GCGCCCCTACCTAGCTGCCT-3′, R: 5′-GGTGGAGACGCGGAGGGAGA-3′. As a negative control for the different pulldowns, absence of GAPDH amplification is shown29.
Human HK3 and KLF5 promoter reporter assays and mutagenesis
Two HK3 promoter regions and one KLF5 promoter region containing the CEBPA binding sites were PCR amplified from genomic DNA of NB4 APL cells using proof reading Pfu DNA polymerase (Promega, Dübendorf, Switzerland) and cloned into the pGL4.10-basic vector (Promega). All mutants were introduced using the QuickChange site-directed mutagenesis system (Agilent, CA, USA) according to the manufacturer's instructions. Promoter transactivation assays were performed as previously described10. Briefly, H1299 cells were transiently transfected with 40 ng of the promoter reporter constructs together with pcDNA3.1 empty or CEBPA expression vector. The promoter activity was measured using the Dual-Luciferase Reporter Plasmid System (Promega) and is shown as relative light units (RLU) relative to pcDNA3.1 control transfected cells.
TaqMan Low Density Array (LDA) and real-time quantitative reverse transcription PCR (qPCR)
RNA extraction, RT-PCR and LDA measurements, as well as data analysis were done as described10,11. Total RNA was extracted using the RNeasy Mini Kit and the RNase-Free DNase Set according to the manufacturer's protocol (Qiagen, Hombrechtikon, Switzerland). Total RNA was reverse transcribed using random primers (Roche Diagnostics) and M-MLV reverse transcriptase (Promega). PCR and fluorescence detection were performed using the ABI PRISM® 7500 Sequence Detection System (Applied Biosystems, Rotkreuz, Switzerland). Data represent log2 expression levels and the measured cycle threshold (Ct) values were normalized to the expression levels of the housekeeping genes HMBS and ABL1.For quantification of KLF5, CEBPA, CEBPE and G-CSFR mRNA in human cells we used Taqman® Gene Expression Assays Hs00156145_m1, Hs00269972_s1, Hs00357657_m1 and Hs00167918_m1, (Applied Biosystems), respectively. HMBS as well as ABL-1 primers and probes have been previously described10,11.
Cell lysate preparation and Western blotting
Whole cell extracts were prepared using RIPA lysis buffer supplemented with 8 M UREA according to the protocol found at www.abcam.com. 40–60 μg total protein was loaded on a 10% or 12% denaturing polyacrylamide gel. Blots were incubated with the primary antibodies in TBS 0.05% Tween-20/5% milk overnight at 4°C, incubated with secondary antimouse or -rabbit antibody at 1:5–10,000 for 1 h at room temperature and analyzed using the Odyssey infrared imaging system detection (Li-Cor Biosciences). Primary antibodies used were anti-CEBPA (Abcam) 1:500 and anti-GAPDH (Millipore) 1:5′000. Secondary antibodies used were goat anti-rabbit IRDye ® 800CW and goat anti-mouse IR Dye ® 680LT (Li-Cor Biosciences).
Transient transfection
HT93 cells were transiently transfected using a cell nucleofector kit (Amaxa Biosystems) according to the manufacturer's instructions. Briefly, the cells were washed in PBS and then resuspended in nucleofector solution to a final concentration of 2 × 106 cells per 100 μl each. Afterwards, the cell suspension was mixed with plasmid DNA pcDNA3.1 either empty, as a negative control, or expressing CEBPA, using the program X-01 and incubated at 37°C and 5% CO2 for 24 h.
Lentivirus preparation and transduction of target cells
pLKO.1-puro lentiviral vectors expressing shRNAs targeting CEBPA (shCEBPA_1: NM_004364.2-171s1c1 or shCEBPA_2: NM_004364.2-1335s1c1) or the control vector (SHC002) were purchased from Sigma-Aldrich. All vectors contain a puromycin antibiotic resistance gene for selection of transduced mammalian cells. Lentivirus production and transduction were done as described10,11. Transduced HT93 and NB4 cells were selected with 1.5 μg/ml puromycin for 4 days and knockdown efficiency was validated by qPCR.
Statistical analysis
Nonparametric Mann-Whitney-U (MWU) tests were applied to compare the difference between two groups using GraphPad Prism 4 software (Graph Pad Software, San Diego, CA). P-values < 0.05 were considered statistically significant. HK3 and KLF5 promoter regions were retrieved from an online database (www.ncbi.nlm.nih.gov), while the putative CEBPA binding sites were predicted by MatInspector (www.genomatix.de).
References
Radomska, H. S. et al. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18, 4301–4314 (1998).
Zhang, D. E. et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 (1997).
Tenen, D. G. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3, 89–101 (2003).
Zeisig, B. B., Kulasekararaj, A. G., Mufti, G. J. & So, C. W. E. SnapShot: Acute myeloid leukemia. Cancer Cell 22, 698–698.e1 (2012).
Paz-Priel, I. & Friedman, A. C/EBPα dysregulation in AML and ALL. Crit Rev Oncog 16, 93–102 (2011).
Pabst, T. & Mueller, B. U. Complexity of CEBPA Dysregulation in Human Acute Myeloid Leukemia. Clin Cancer Res 15, 5303–5307 (2009).
Gombart, A. F. et al. Mutations in the Gene Encoding the Transcription Factor CCAAT/Enhancer Binding Protein A in Myelodysplastic Syndromes and Acute Myeloid Leukemias. Blood 99, 1332–1340 (2002).
Martelli, M. P., Sportoletti, P., Tiacci, E., Martelli, M. F. & Falini, B. Mutational landscape of AML with normal cytogenetics: biological and clinical implications. Blood Rev. 27, 13–22 (2013).
Figueroa, M. E. et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood 114, 3448–3458 (2009).
Federzoni, E. A. et al. PU.1 is linking the glycolytic enzyme HK3 in neutrophil differentiation and survival of APL cells. Blood 119, 4963–4970 (2012).
Humbert, M. et al. Deregulated expression of Kruppel-like factors in acute myeloid leukemia. Leuk. Res. 35, 909–913 (2011).
Rijksen, G, S. G., Rijksen, G., Staal, G. E., Beks, P. J. et al. Compartmentation of hexokinase in human blood cells. Characterization of soluble and particulate enzymes. Biochim. Biophys. Acta 719, 431–7 (1982).
McConnell, B. B. & Yang, V. W. Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 90, 1337–1381 (2010).
Diakiw, S. M. et al. The granulocyte-associated transcription factor Krüppel-like factor 5 is silenced by hypermethylation in acute myeloid leukemia. Leuk. Res. 36, 110–116 (2012).
Pulikkan, J. A. et al. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood 116, 5638–5649 (2010).
Taskesen, E. et al. Prognostic impact, concurrent genetic mutations and gene expression features of AML with CEBPA mutations in a cohort of 1182 cytogenetically normal AML patients: further evidence for CEBPA double mutant AML as a distinctive disease entity. Blood 117, 2469–2475 (2011).
D'Alo', F. et al. The amino terminal and E2F interaction domains are critical for C/EBPα-mediated induction of granulopoietic development of hematopoietic cells. Blood 102, 3163–3171 (2003).
Rosenbauer, F. & Tenen, D. G. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7, 105–117 (2007).
Nerlov, C. C/EBPα mutations in acute myeloid leukaemias. Nature Reviews Cancer 4, 394–400 (2004).
Schlenk, R. F. et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 358, 1909–1918 (2008).
Yang, J. et al. Metabolic response of mice to a postnatal ablation of CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 280, 38689–38699 (2005).
Bereshchenko, O. et al. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 16, 390–400 (2009).
Kirstetter, P. et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 13, 299–310 (2008).
Dong, J.-T. & Chen, C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 66, 2691–2706 (2009).
Karp, J. E. & Rassool, F. V. KLFs and ATRA-induced differentiation: New pathways for exploitation. Leukemia Research 35, 846–847 (2011).
Diakiw, S. M. et al. Methylation of KLF5 contributes to reduced expression in acute myeloid leukaemia and is associated with poor overall survival. Br. J. Haematol. 161, 884–888 (2013).
Chen, C. et al. Regulation of KLF5 involves the Sp1 transcription factor in human epithelial cells. Gene 330, 133–142 (2004).
López-Rodríguez, C., Botella, L. & Corbí, A. L. CCAAT-enhancer-binding proteins (C/EBP) regulate the tissue specific activity of the CD11c integrin gene promoter through functional interactions with Sp1 proteins. J. Biol. Chem. 272, 29120–29126 (1997).
Kaeser, M. D. & Iggo, R. D. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 95–100 (2002).
Acknowledgements
We thank D. Shan for excellent technical support and Dr. Diogo F.T. Veiga for his help in analyzing the public available AML patient data. This study was supported by grants from the Swiss National Science Foundation 31003A_129702 (to M.F.F. and M.P.T.) and 31003A_143739 (to M.P.T.), the Marlies-Schwegler Foundation, the Ursula-Hecht-Foundation for Leukemia Research and the Bernese Foundation of Cancer Research (to M.F.F.), the Werner and Hedy Berger-Janser Foundation of Cancer Research (to M.F.F. and M.P.T.), the Bern University Research Foundation (to M.P.T.) and the Joyce Klein Stock Gift and NIH R01HL091219 (to B.E.T.).
Author information
Authors and Affiliations
Contributions
E.A.F. and M.H. performed the experimental research, analyzed the data and drafted the article. G.B. provided essential CEBPA reagents and revised the article. M.F.F. and B.E.T. instigated the experimental design and revised the drafted article. M.P.T. designed the project, wrote the paper and gave final approval of the submitted manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Federzoni, E., Humbert, M., Torbett, B. et al. CEBPA-dependent HK3 and KLF5 expression in primary AML and during AML differentiation. Sci Rep 4, 4261 (2014). https://doi.org/10.1038/srep04261
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04261
This article is cited by
-
Mouse strain-specific polymorphic provirus functions as cis-regulatory element leading to epigenomic and transcriptomic variations
Nature Communications (2021)
-
Venetoclax and pegcrisantaspase for complex karyotype acute myeloid leukemia
Leukemia (2021)
-
HK3 overexpression associated with epithelial-mesenchymal transition in colorectal cancer
BMC Genomics (2018)
-
C/EBPα in normal and malignant myelopoiesis
International Journal of Hematology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.