Abstract
HIV has become one of the most devastating pathogens in human history. Despite fast progress in HIV-related basic research, antiretroviral therapy (ART) remains the most effective method to save AIDS patients' lives. Unfortunately, ART cannot be universally accessed, especially in developing countries, due to the lack of effective treatment monitoring diagnostics. Here, we present an inexpensive, rapid and portable micro-a-fluidic platform, which can streamline the process of an enzyme-linked immunosorbent assay (ELISA) in a fully automated manner for CD4 cell count. The micro-a-fluidic CD4 cell count is achieved by eliminating operational fluid flow via “moving the substrate”, as opposed to “flowing liquid” in traditional ELISA or microfluidic methods. This is the first demonstration of capturing and detecting cells from unprocessed whole blood using the enzyme-linked immunosorbent assay (ELISA) in a microfluidic channel. Combined with cell phone imaging, the presented micro-a-fluidic ELISA platform holds great promise for offering rapid CD4 cell count to scale up much needed ART in resource-constrained settings. The developed system can be extended to multiple areas for ELISA-related assays.
Similar content being viewed by others
Introduction
HIV has significantly impacted basic and clinical research for better patient care by causing more than 25 million deaths and 35.3 million people infected worldwide1. Although antiretroviral therapy (ART) is effective in saving AIDS patients' lives, the implementation of ART worldwide has been drastically hampered by the lack of treatment monitoring diagnostics. According to WHO 2013 guidelines, the coverage of ART is still only 34% in low- and middle-income countries1, despite ART being affordable or even free. The fundamental challenge is that AIDS patients on ART need to be closely monitored for drug adherence and efficacy, but existing viral load and CD4 cell count assays generally remain costly and technologically complex2. In addition, these monitoring diagnostics cannot be implemented in rural areas where basic laboratory infrastructure and trained operators are lacking. Thus, a point-of-care (POC) diagnostic tool is highly desirable to deliver treatment monitoring to AIDS patients wherever and whenever needed.
Microfluidics have been widely utilized to develop POC diagnostics and thus to address global healthcare issues, including HIV3,4. However, the requirement for precise control of fluid flow often dictates the need for bulky, expensive instruments and well-trained operators, which significantly impedes the translation of microfluidic technologies into clinical practice5,6. Researchers, for example, need to achieve fine manipulation of different fluids in terms of when, where and how much for accurate measurements in a multiple-step biological reaction process such as ELISA. The hidden cost and complexity associated with precise fluid flow constitute a critical barrier for performing microfluidic-based diagnosis at the POC. The complex designs required to precisely control fluid flow in microfluidic devices (e.g., addition of valves) are generally not yet amenable to automation for POC testing7. Moreover, the generation of air bubbles in flow-based microfluidics devices due to switching of liquid reagents can potentially block microchannels, thus compromising or even disabling targeted biological reactions. Hence, the development of flow-free, bubble-free, portable devices is a pre-requisite to perform simple, rapid and inexpensive ART monitoring at the POC.
Herein, we developed a micro-a-fluidic ELISA (m-ELISA) to rapidly measure CD4 cell count in an automated manner. This process is initiated by m-ELISA and concluded with a cell phone-based colorimetric detection (Fig. 1a). In the micro-a-fluidic system, fluids are stationary, while the captured CD4+ T lymphocytes are mobile. We demonstrated the unique capability of m-ELISA to capture and detect cells from whole blood without sample processing (e.g., cell fixing), which cannot be achieved in traditional flow-based ELISA due to high shear stress. The m-ELISA detected down to 30 CD4+ T lymphocytes per reaction in a total assay time of 9 minutes. Among the unprocessed whole blood samples of 35 patients, the m-ELISA assay measured CD4+ T cell counts with an accuracy of 97% at the clinical cutoff (e.g., 350 cells/μL) recommended by the World Health Organization (WHO)20 for prioritizing ART, thus offering a “plug-and-play” solution to the scale-up of ART in AIDS patients in resource-constrained settings.
Design of m-ELISA for CD4+ T lymphocyte count.
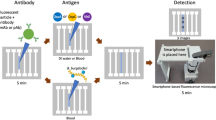
(a) Overview of the m-ELISA based chip platform. The testing process can be divided into three steps consisting of sample collection, rapid cell-based m-ELISA and cell phone based colorimetric detection. A droplet from a finger prick can be collected and loaded onto a micro-a-fluidic chip along with antibody-functionalized magnetic beads. The micro-a-fluidic chip is placed on a permanent magnet, which is fixed on a motorized stage. With the aid of a software program, the stage is used to control and complete the entire process of ELISA in an automated manner. As a result, the color development on-chip is imaged using a cell phone and the result is analyzed using an integrated mobile application. (b) Design of a micro-a-fluidic chip and cell-based CD4 ELISA. The chip is designed to contain five aqueous circular chambers (i, ii, iii and two wash chambers containing phosphate buffered saline plus 0.5% tween 20 (PBST)) and five elliptical chambers containing mineral oil for physical separation of the hydrophilic circular chamber liquids. In chamber i, functionalized magnetic beads conjugated with anti-CD4 antibody are used to selectively capture cells (CD4+ T lymphocytes and monocytes) expressing CD4 molecules on the surface. After a washing step in the intermediate chamber, the magnetic beads with captured CD4+ cells are moved to chamber ii and tagged with anti-CD3 antibody, which is conjugated with horseradish peroxidase (HRP). After another wash, the captured CD4+ T lymphocytes enable color development due to digestion of 3,3′,5,5′-Tetramethylbenzidine (TMB) by HRP in chamber iii. The photograph in (a) was taken by ShuQi Wang.
Results
Working principle of micro-a-fluidic ELISA
The m-ELISA eliminates fluid flow by “moving the substrate” through different aqueous phases, in contrast to “flowing liquid” through a microwell or microchannel as performed in the traditional flow-based ELISA. The temporal processes such as sample incubation, washing, enzyme labeling and colorimetrical detection were performed spatially via manipulating magnetic beads in a micro-a-fluidic device (Fig. 1b). Five circular chambers were separately pre-loaded with whole blood plus antibody-functionalized magnetic bead, wash buffer, horseradish peroxidase (HRP)-conjugated antibody, wash buffer and 3,3′,5,5′-tetramethylbenzidine (TMB) solution. The 5 elliptical chambers located between the circular chambers contained mineral oil for physical phase separation of reagents. Mineral oil physically separated the aqueous solutions within the circular chambers while still allowing for the transmission of the CD4+ cell-magnetic bead complexes. In the first circular chamber, magnetic beads functionalized with anti-CD4 antibody were used to capture CD4+ cells from whole blood. The magnetic beads carrying captured CD4+ cells were then washed in the second circular chamber to remove unbound, non-specifically attached cells. In the third circular chamber, HRP-conjugated anti-CD3 antibody was tagged onto the captured CD4+ cells. After another wash in the fourth chamber, the HRP-tagged CD4+ cells induced the oxidation of TMB and generated blue color in the fifth chamber. The color intensity was quantified using a smart phone with an integrated mobile application8. The entire ELISA process was automated to complete within 9 minutes using an in-house-developed portable magnetic actuator (Supplementary Fig. 1).
Characterization of CD4+ cell capture on magnetic beads
Characterization of the capture of CD4+ cells on magnetic beads that were functionalized with anti-CD4 antibody, was carried out using immunostaining and scanning electron microscopy (SEM). In Fig. 2a, a CD4+ T lymphocyte (both CD3 and CD4 positive) was captured on a magnetic bead and stained using Alexa 488-labeled anti-CD4 antibody emitting green fluorescence. Since monocytes express CD4 molecules (but not CD3 molecules), monocytes can also be captured on the beads. To differentiate the emission of CD4+ T lymphocytes from monocytes, we used Alexa 647-labeled anti-CD3 antibody (Fig. 2a). This antibody does not stain monocytes, but binds to CD4+ T lymphocytes. The overlaid fluorescence micrographs showing the emission of Alexa 488 and 647, as well as the bright field and SEM images confirm the capture of CD4+ T lymphocytes on magnetic beads (Fig. 2a). The capture efficiency of CD4+ T lymphocytes on magnetic beads was analyzed also by means of flow cytometry. The results showed that the capture efficiency was 68 ± 4% over incubation times varying from 1 to 10 min and there was no statistically significant difference in capture efficiency among the varying incubation times (p > 0.05, n = 3) (Fig. 2b). These data with confidence indicate that CD4+ T lymphocytes were captured on magnetic beads by anti-CD4 antibodies in a few minutes. In addition, we evaluated the capture efficiency over the decreasing concentrations of CD4+ T lymphocytes (Fig. 2c). The highest capture efficiency was 69 ± 3.6% for 1,200 cells/μL; the lowest capture efficiency was 60.3 ± 6.0% for 50 cells/μL. One-way ANOVA analysis indicated that there was no statistically significant difference in capture efficiency over the concentrations of CD4+ T lymphocytes ranging from 50–1200 cells/μL.
Characterization of m-ELISA for CD4 cell count.
(a) Capture of CD4+ T lymphocytes (red arrow) on magnetic beads (blue arrow). Magnetic beads were incubated with whole blood and the captured T lymphocyte was stained with anti-CD4 Alexa Fluor 488 and anti-CD3 Alexa Fluor 647. The fluorescent images of Fluor 488, Alexa Fluor 647, bright-field, as well as the overlaid image (by ImageJ software) demonstrate capture of CD4+ T lymphocytes by magnetic beads. The scanning electron microscope (SEM) image validates the capture of CD4+ T lymphocyte on magnetic beads. The scale bar is 10 μm. (b) Efficiency of magnetic beads for capturing CD4+ T lymphocytes at varying incubation times. 25 μL of functionalized magnetic beads (10 mg/mL) were incubated with whole blood containing 1,000 cells/μL CD4+ T lymphocyte. The unbound T lymphocytes in the supernatant were quantified by flow cytometry, which was used to calculate the capture efficiency. One-way ANOVA was performed to analyze and no statistical significance (p > 0.05) was found. (c) Efficiency of magnetic beads for capturing CD4+ T lymphocytes at varying concentrations of CD4+ T lymphocytes. The same procedure as mentioned in (b) was used to calculate the capture efficiency. One-way ANOVA analysis indicated that there was no significant difference in capture efficiency (p > 0.05). (d) Optimization of HRP-antibody in m-ELISA. The variable, HRP-conjugated antibody was diluted at different concentrations and tested with plasma samples, which served as negative control. 1:80,000 was chosen for the following experiments since this concentration was the highest concentration of HRP-antibody that avoided false positive results. A mobile application was used to extract red (R) pixel values from the color development region on-chip and normalized them according to the background as shown in the Y-axis. One-way ANOVA was performed to analyze the color intensity caused by excessive HRP (n = 3, * indicates the statistical significance, p < 0.05). (e) Whole blood from a healthy blood sample was serially diluted (undiluted, 1:3, 1:9, 1:27 and 1:81) used to construct the standard curve with different 3, 3′, 5, 5′ Tetramethylbenzidine (TMB) incubation periods. Each data point was performed in triplicates and the standard deviation was shown.
Development of the protocol of micro-a-fluidic ELISA for CD4 cell count
We further developed a rapid protocol of m-ELISA for quantification of CD4+ T lymphocytes. To differentiate CD4+ T lymphocytes from monocytes, we used anti-CD3 antibody to eliminate potential contaminant of monocytes, which also have CD4 molecules on the cell membrane. We first evaluated various concentrations of HRP-conjugated anti-CD3 antibody (1:1,000, 1:10,000, 1:80,000, 1:100,000 and 1:1,000,000 dilutions from stock concentration) in m-ELISA to reduce the background readout. In these experiments, a plasma sample was used as a negative control, where a whole blood sample was used as a positive control for CD4 cell count. The results showed that 1:80,000 dilution of anti-CD3-HRP antibody only developed slightly, close to negligible blue color with a normalized R-value of 0.03 (Fig. 2d). We chose the 1:80,000 dilution of anti-CD3-HRP antibody as a working concentration (0.52 ng/mL) in the following experiments, as it also elicited sufficient color development in positive samples as opposed to the dilutions of 1:100,000 and 1:1,000,000. We then evaluated the impact of TMB incubation time on color development (Fig. 2e). The standard curve with a 5 min TMB incubation gave a better fit (R2 = 0.98) compared to other incubation durations evaluated. Comparatively, longer incubation duration (10 min) led to saturation of color development and shorter incubation (2 min) did not elicit sufficient color development – neither of which would give accurate ELISA quantification.
Modeling of magnetic forces in micro-a-fluidic devices
To better understand the manipulation of magnetic beads in a micro-a-fluidic device, we developed a computational model to evaluate magnetic forces, fluidic drag forces and interfacial forces on beads in a magnetic field (Fig. 3). Based on the computational model, we evaluated two magnetic forces: (i) magnetic forces that vertically pull magnetic beads towards the base of the chamber (Fig. 3a) and (ii) magnetic forces that pull beads parallel to the axis of the magnet motion (Fig. 3b). Since the magnetic susceptibility of PMMA, water and oil used in our experimental setup is much smaller (χ ≪ 1) than that of magnetic beads (χ ≫ 1)9, the contribution to magnetic field is negligible compared to magnetic beads and thus we assume the medium surrounding the magnet is uniform. The results showed that as the distance between magnetic beads and the cylindrical magnet underneath decreased, both vertical and horizontal magnetic forces increased. Theoretically, if magnetic beads were located above a critical height (h) from the surface of magnets, magnetic forces exerted on beads would not be large enough to overcome Brownian forces. To predict this limitation of our design, we evaluated this height as a function of the magnetic bead size. Critical height (h) for magnetic particles is calculated by Eq. 1–4 in the Supplementary Information and plotted as a function of radius of the bead in Fig. 3c. Results showed that with current settings and parameters of the experimental setup, Brownian motion is not effective in our experiments. Next, we calculated the capillary pressure difference sustained across the interface between mineral oil and PBS and compared them with horizontal magnetic forces (Fig. 3d). Since the shape of oil on-chip largely depends on the elliptical chamber as shown in Fig. 1, although the elliptical shape of oil changed slightly after testing, we used the shape of ellipsoid (b and a are the major and minor radii) to calculate capillary pressure. Then capillary pressure difference can be written  , where mean curvature, H(t) = ab/(b2cos2t + a2sin2t)3/2. Here, t = 0 corresponds to points where ellipsoidal chambers are intersected with the major horizontal path of the magnetic beads. Then, capillary pressure difference, ΔP = −2γa/b2, where γ is surface tension of mineral oil against PBS (γ = 50 mN/m). In Fig. 3d, ΔP (Pa) are plotted as a function of b/a (dashed blue line) and corresponding shapes of chambers are also plotted in the figure. Magnetic forces (z = 2 mm) per cross-section area of magnetic beads are plotted as a function of r/R (solid red line). Results showed that as b/a ratio decreases (or ellipsoidal chambers become more circular) capillary pressure difference increases. We also plotted magnetic flux density (contours) and unit magnetic forces (red arrows) when a magnet moves horizontally from left to right (Fig. 3e, Supplementary Fig. 2a).
, where mean curvature, H(t) = ab/(b2cos2t + a2sin2t)3/2. Here, t = 0 corresponds to points where ellipsoidal chambers are intersected with the major horizontal path of the magnetic beads. Then, capillary pressure difference, ΔP = −2γa/b2, where γ is surface tension of mineral oil against PBS (γ = 50 mN/m). In Fig. 3d, ΔP (Pa) are plotted as a function of b/a (dashed blue line) and corresponding shapes of chambers are also plotted in the figure. Magnetic forces (z = 2 mm) per cross-section area of magnetic beads are plotted as a function of r/R (solid red line). Results showed that as b/a ratio decreases (or ellipsoidal chambers become more circular) capillary pressure difference increases. We also plotted magnetic flux density (contours) and unit magnetic forces (red arrows) when a magnet moves horizontally from left to right (Fig. 3e, Supplementary Fig. 2a).
Computational modeling of driving forces exerted on magnetic beads.
(a) Magnetic forces on a single magnetic bead in z-direction for a range of vertical distance, z = 1, 2 and 3 mm. (b) Magnetic forces on a single magnetic bead in r-direction. (c) Critical height (h), i.e., the location where magnetic forces exerted on beads is comparable to Brownian forces, is calculated as a function of magnetic bead size (Rp) using Eq. 1–4 given in the supplementary information. Brownian motion of magnetic particles is not desirable because such motion would limit the bead transport. Results showed that Brownian forces are not effective in our experiments with the current settings and dimensions of the setup. (d) Capillary pressure difference sustained across the interface between mineral oil and PBS (dashed blue line) and magnetic forces (z = 2 mm) per cross-section area of magnetic beads (solid red line). (e) Simulation results of magnetic flux density (contours) and unit magnetic force along the horizontal plane (arrows) when a magnet moves horizontally from left to right. Results are plotted for a range of time points (top-to-bottom, left-to-right). Full results are given in the supplementary information. Dashed lines are the circular edges of the cylindrical magnet (top view).
Validation of micro-a-fluidic ELISA for CD4 cell count
Finally, we demonstrated that the accurate CD4+ T lymphocyte counts by means of m-ELISA was suitable for monitoring of antiretroviral therapy in AIDS patients. We tested 17 AIDS patients and 18 transplantation patients with artificially suppressed immune systems for CD4+ T lymphocyte counts. Compared to the gold standard for CD4 cell count, i.e., flow cytometry, m-ELISA results showed a linearity of 0.86 (R2) and 0.90 (R2) in AIDS patients and transplantation patients, respectively (Fig. 4a, b). In AIDS patients without therapeutic intervention, the CD4 cell count generally tends to be low. Since the samples were collected from AIDS patients who were on treatment, the CD4 cell count of most patients exceeded the clinical cutoff (350 cells/μL) recommended by the WHO for prioritizing ART in resource-constrained settings20. The CD4 cell count with m-ELISA ranged from 237 to 1,446 cells/μL in AIDS patients and from 146 to 1,237 cells/μL in transplantation patients. Further, we evaluated the discrepancy in CD4 cell count in both populations measured by m-ELISA and flow cytometry (Fig. 4c, d). Of 18 transplantation patients, only 1 sample had CD4 cell count exceeding ±1.96 S.D (Fig. 4e). Among 35 patients, m-ELISA had an accuracy of 97.1% in identifying patients with CD4 cell count above or below 350 cells/μL. These data demonstrate that the m-ELISA platform can reliably report CD4 cell count for potential use in monitoring ART in AIDS patients in resource-constrained settings.
Clinical validation of micro-a-fluidic ELISA for CD4+ T lymphocyte count.
(a) and (b) Correlation of flow cytometry with micro-a-fluidic ELISA for CD4+ T lymphocyte count in 17 AIDS patient samples and 18 immunosuppressed patient samples, respectively. (c) and (d) Bland-Altman analysis for the difference between flow cytometry and micro-a-fluidic ELISA for CD4+ T lymphocyte count in 17 AIDS patient samples and 18 immunosuppressed patient samples, respectively. The 95% confidence interval on the mean is shown on the graphs. (e) Accuracy of CD4+ T lymphocyte micro-a-fluidic ELISA at the WHO clinical cutoff of 350 cells/μL for prioritizing ART. The samples were defined as positive once flow cytometry count was below 350 cells/μL, otherwise negative.
Discussion
To the best of our knowledge, this report is the first demonstration that cells can be reliably detected in ELISA in such a non-flow manner. Because there is no fluid flow, cells can be captured on the surfaces of magnetic beads without experiencing high shear stress, which occurs in a conventional flow-based system. Due to high shear stress generated in flow-based ELISA (i.e., multiple wash steps at high speed), there is no report on the detection of cells attached to a solid phase in a microfluidic channel. It should be noted that cells could be physically blocked and detected in filter-based microfluidic devices, which allows for stringent washing and withstanding to resultant high shear stress10. However, this flow-based ELISA strategy as well as the use of fluorescence detection is not suitable for POC testing according to the ASSURED criteria (i.e., affordable, sensitive, specific, user-friendly, rapid, equipment-free and deliverable)11. Although shear stress can be significantly lowered at slow flow rates, the assay time would be inevitably prolonged, which is not ideal for POC testing. The micro-a-fluidic system was designed to accommodate POC testing with unique features such as a high degree of automation, short assay turnaround time and cell phone-based imaging/readout. A brief comparison of these aspects among the flow-based ELISA and the m-ELISA is presented in Supplementary Table 1.
With its simplicity and rapidity, this micro-a-fluidic CD4+ T lymphocyte cell counting method can considerably facilitate the expansion of ART in resource-constrained settings. In 2012, the coverage of ART in low- and middle-income countries was approximately 34% despite free provision of antiretroviral drugs in most developing countries1, due mainly to the lack of treatment monitoring tools2. Until now, a variety of CD4 assays have been developed to replace flow cytometry, i.e., the “gold standard rapid CD4 cell count platform”. On one hand, simplified flow cytometers such as EasyCD412, CyFlow13 and Pan-leucogating14 have been developed to reduce the instrumentation cost. Furthermore, manual CD4 assays such as Dynabeads and Cytosphere have been developed to eliminate the need for instrumentation, which are more suitable for de-centralized laboratories. On the other hand, microfluidic-based CD4 cell counting technologies have moved rapidly forward, leveraging immune-based cell capture from whole blood in microchannels10,15,16,17. Nevertheless, existing cell detection and counting methods still have challenges such as high cost, technological complexity, labor-intensiveness and most importantly inherent “liquid flow” for POC testing.
In conclusion, a rapid, automated m-ELISA platform was developed for POC testing in resource-constrained settings with a demonstration of an unmet need for rapid CD4+ T lymphocyte cell count. This POC testing platform holds great potential to offer much needed treatment monitoring services and significantly impacts the scale-up of ART missioned by the WHO in resource-constrained settings. In the future, this micro-a-fluidic platform can be broadly applied to the detection of other types of cells for early diagnosis and/or treatment monitoring of various diseases including cancer.
Methods
Device fabrication
The m-ELISA chips for CD4+ cell counting were assembled using a non-lithographic technique as we previously reported18,19,21,22. To assemble this device, poly(methyl methacrylate) (PMMA) (McMaster-Carr, Atlanta, GA) and double-side adhesive film (DSA) (50 μm thick, iTapestore, Scotch Plains, NJ) were cut using a LASER cutter (Versa Laser ™, Scottsdale AZ). The device dimensions measured 43 mm × 63 mm × 6 mm. Three layers of PMMA of 2 different thicknesses (1.5 mm and 3.175 mm) were used; the top and bottom layer of PMMA were 1.5 mm thick and the middle layer of PMMA containing micro-chambers was 3.175 mm thick. The middle PMMA layer was designed to contain 5 circular chambers (chambers i, ii and iii, as well as two wash chambers; chamber i having a radius of 4.5 mm and the rest circular chambers having with a radius of 3.5 mm) to contain ELISA reagents. These circular chambers were connected by 4 elliptical chambers (with major and minor axes of 13.5 mm and 3.8 mm, respectively), containing mineral oil (Sigma-Aldrich, St. Louis, MO) to separate ELISA reagents (Fig. 1b). After chamber iii, there was another elliptical chamber (with major and minor axes of 13.5 mm and 6.5 mm, respectively) containing mineral oil. The top PMMA layer contained a single opening (0.4 mm radius) to load sample/reagent in the circular chambers and two congruent openings for loading mineral oil in the elliptical chambers, one to load and the other to allow for the escape of air. The bottom PMMA layer had no openings. Two layers of DSA, with the same design as the middle PMMA layer, were used to assemble the micro-a-fluidic chips.
Functionalization of magnetic beads with primary anti-CD4 antibodies
Protein G (PG) coated magnetic beads with a mean diameter of 2 μm (New England Biolabs, Ipswich, MA) were used to serve as a solid phase to immobilize anti-CD4 monoclonal capture antibodies. In brief, the entire content of the Protein-G bead vial (2.16 × 10∧6 particles/ul, 1 mL) was removed by pipette to a 15-mL falcon tube (BD Bioscience, Woburn, MA). 3 mL of Dulbecco's phosphate buffered saline (DPBS) (Invitrogen, Carlsbad, CA) was added on the PG beads. The falcon tube was applied onto a magnetic stand for 1 minute to allow the PG beads to aggregate on the falcon tube wall directly adjacent to the magnetic stand. The supernatant was removed by pipetting and replaced with 4 mL DPBS, in order to wash the PG beads. After gentle mixing for 1 min, the supernatant was once again removed and the washing step was repeated 3 times. For CD4+ cell capturing, 1 mL of 5 μg/mL mouse monoclonal anti-CD4 antibody (Abcam, Cambridge, MA) was conjugated to the PG beads. The antibody-magnetic bead solution was incubated overnight at 4°C on a rotator for conjugation. After incubation, the PG beads were washed three times as mentioned above to remove unbound anti-CD4 antibodies. The functionalized PG magnetic beads were then blocked with 3% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) for 1 hour at room temperature. After another three-time wash using 1 mL of DPBS, the magnetic beads were re-suspended in 1 mL of DPBS and stored at 4°C prior to use.
HRP conjugation of secondary anti-CD3 antibodies
Rabbit polyclonal secondary anti-CD3 antibody (0.2 mg/mL, Abcam, Cambridge, MA) was conjugated with horseradish peroxidase (HRP) using an EasyLink HRP Conjugation Kit (3 × 100 μg Biotin, Abcam, Cambridge, MA). According to the manufacturer's instructions, 10 μL of EL-Modifier reagent was mixed gently with 100 μl of anti-CD3 antibody (1:80,000 dilution) in a 1.5 mL Eppendorf tube. After mixing, 100 μL of EL-HRP mix was added. The resultant mixture was incubated overnight in dark at ambient temperature. Following this, 10 μL of EL-Quencher reagent was added to stop the conjugation process. The working concentration of anti-CD3 antibody was experimentally determined to be 1:80,000 diluted from the conjugated solution, at which there was negligible color in the serum samples (serving as negative controls) while still leading to strong color in positive standards.
Immunofluorescence staining of CD4+ T lymphocytes captured on magnetic beads
100 μL of 1.5 μg/mL anti-CD4 Alexa Fluor 488 (BioLegend, San Diego, CA) was added to 100 μL of whole blood (Massachusetts General Hospital, Boston, MA) in an Eppendorf tube and incubated for 40 minutes in darkness at ambient temperature. After incubation in order to wash away the excess antibody, 200 μL of 0.05% tween-20 washing buffer (DPBST), was added. The resultant solution was mixed gently and centrifuged for 10 seconds at 6000 RPM (Labnet Spectrafuge Mini Centrifuge, Korea). The supernatant was then removed and the pelleted cells were washed twice through the addition of DPBS and subsequent removal of supernatant. Next, 100 μL of 6 μg/mL Anti-CD3 Alexa Fluor 647 was added to the Eppendorf tube and incubated for 40 minutes in darkness at ambient temperature. Again, the washing step was preformed 3 times as mentioned above. Afterwards, 20 μL of anti-CD4 antibody coated magnetic beads were added and incubated for 10 minutes at room temperature in darkness. After incubation, magnetic beads with captured CD4+ cells were collected using a magnetic stand and were washed with 200 μL of DPBST twice. 10 μL of DPBS was added to re-suspend the magnetic beads with captured CD4+ cells and 1 μL of the mixture was mounted to a glass slide for fluorescence imaging. Magnetic beads conjugated with CD4+ T lymphocytes were mounted to a glass slide for fluorescence and bright-field imaging.
Scanning Electron Microscopy (SEM) Imaging
The cells were fixed and dehydrated using ethanol solution. The cells were then dried of ethanol on coverslips and mounted onto aluminum SEM sample holders (Ted Pella Inc., Redding, CA) using conductive carbon tape (Ted Pella Inc., Redding, CA). The mounted samples were coated with Platinum/Palladium at 40 mA for 90 seconds (Cressington Scientific Instruments Ltd., Watford, England) in an Argon gas purged chamber. After coating, samples were imaged using field emission SEM (Ultra 55, Carl Zeiss Micro Imaging LLC, Thornwood, NY) under high vacuum mode using in-lens detector. The samples were imaged at 10,000×–30,000× magnifications at 5 kV accelerating voltage and <5 mm working distance.
Reagent loading
HRP-conjugated anti-CD3 rabbit polyclonal antibody (1:800,000 dilution), one-Step ultra 3,3′,5,5′-tetramethylbenzidine (TMB) (Thermo Scientific Inc., Waltham, MA) and 100 μL of PBST was pipetted into chambers ii, iii and the unlabelled smaller circular chambers, respectively (Fig. 1b). These small volumes of reagents filled up the circular chamber volume without leaking into adjacent elliptical chambers due to high surface tension from the interface between the liquid reagents and the inner surface of the micro-a-fluidic chip. After these reagents were input, approximately 213 μL of mineral oil was injected to fill the last elliptical chamber. And 125 μL of mineral oil was loaded into other elliptical chambers, except for the first elliptical chamber (between chamber i and the first wash chamber). Next, 25 μL of the magnetic beads conjugated with mouse monoclonal anti-CD4 capture antibody and 10 μL of blood sample was loaded into the sample chamber i. DPBST was then added to fill the remaining volume of chamber i. Lastly, 125 μL of mineral oil was loaded into the remaining elliptical chamber.
m-ELISA by magnetic actuation
Magnetic force was utilized to mechanically actuate the movement of magnetic beads through all the chambers by use of a magnetic stage that was fully automated via a program and the total assay time was 9 minutes. The loaded micro-a-fluidic chip was placed in an appropriate slot of the portable instrument (Supplementary Fig. 2). The actuation of permanent magnet was automated (Video 1, Supplementary File 2) using an in-house developed program.
Upon the placement of the micro-a-fluidic chip on the instrument, a permanent magnet (1/4″ dia. × 1″ thick, K&J Magnetics, Inc.) underneath chamber i settled the magnetic beads with captured CD4+ T lymphocytes via antibody-antigen interaction. The PG magnetic beads with captured CD4+ T lymphocytes were moved to the next circular chamber, crossing the oil barrier in the elliptical chamber, via the NdFeB magnet placed on a linear X-direction stage. In this chamber, bead washing was performed with wash buffer by actuating the conjugated beads back and forth within the wash chamber. The back and forth motion of the magnet resulted in collection of residual magnetic beads left in the previous elliptical oil chamber. After mixing for 1 minute in the wash chamber, the beads were further actuated to chamber ii containing the HRP-conjugated anti-CD3 secondary antibody. While mixing for 1 minute in this chamber, captured CD4+ T lymphocytes interacted with the anti-CD3 antibody, forming an anti-CD4 antibody/CD4+ T lymphocyte/anti-CD3 antibody sandwich structure. The magnetic beads with the immune complex were then moved to the second wash chamber. Following another washing step, the magnetic beads with the immune complex were moved to chamber iii containing TMB and accordingly mixed for 6 minutes. Due to the presence of HRP-antibody captured on the magnetic beads, the substrate TMB was digested and a blue color was developed. Thus, the total assay time was 9 minutes. The magnetic beads were removed from chamber iii into the last elliptical oil chamber and actuated back and forth for an additional minute to attract and remove any residual beads left in chamber iii. While bead mixing, the blue solution in chamber iii was mixed manually using a pipet to achieve a homogeneous color density.
Imaging and data analysis using a cell phone
The m-ELISA-based chip with homogeneous blue color in chamber iii was immediately placed onto a white LED-illuminated translucent white acrylic plexiglass (McMaster-Carr, Atlanta, GA) sheet inside a black plastic box (Supplementary Fig. 3). The black box was used to provide an isolated light environment, where light variation in ambient environment is reduced. The internal LED lighting setup was to achieve even background during imaging. On top of the black box, there was an opening where a cell phone with a built-in camera was used to image the color intensity. To facilitate image processing, we developed a desktop application that automatically selects the region for colorimetric analysis, calculates R values and normalizes R values according to the background in a batch-processing format (Supplementary Fig. 4). The normalized red pixel value of negative control (plasma) as a result of 1:80,000 HRP antibody was used as a basis to determine the sensitivity (mean ±2 standard deviation). In addition, we developed a mobile application that has the same analysis functionality as the desktop application but also constructs the standard curve and reports the quantification results (Supplementary Fig. 5).
Flow cytometry
The CD4 cell count of clinical samples was measured using the gold standard – flow cytometry. Before measurement, the FACSCalibur system (Beckton Dickinson, San Jose, CA) was calibrated using Becton Dickinson BD CaliBRITE™ Beads. Becton Dickinson Multi-Check CD4 Low Control and Multi-Check Control were used in parallel with clinical samples to ensure accuracy and reliability of flow cytometry. For each measurement, 20 μL of antibody mixture anti-CD3/CD8/CD45/CD4 antibodies and 50 μL blood or control were added to Trucount™ Tubes. After incubation for 15 minutes at room temperature, red blood cells were lysed in 450 μL of 1× FACS Lysing Solution. After another 15 min-incubation, the mixture was loaded to the flow cytometer for CD4 cell count using Becton Dickinson MultiSET™ software. For assessing the capture efficiency of CD4 T+ T lymphocytes to magnetic beads, it was calculated based on the following equation.

Clinical testing
With the approval of the Institutional Review Board (IRB) (protocol: 2009P0001790), 17 discarded anonymous and de-identified HIV-infected whole blood patient samples were collected from the Massachusetts General Hospital (MGH). In addition, 18 discarded immunosuppressed patient samples were tested using the micro-a-fluidic CD4 assay. These clinical samples were tested in duplicates using the micro-a-fluidic ELISA. In parallel, a whole blood sample from a healthy blood donor was serially diluted and tested (including undiluted, 1:3, 1:9, 1:27 and 1:81) to construct a standard curve for cell quantification. The CD4 cell count of the undiluted whole blood sample was analyzed using flow cytometry. The normalized red pixel numbers of the clinical samples were interpolated with the logarithmic fit equation of the standard curve and CD4+ T lymphocyte count was calculated.
Statistics analysis
In this study, we used MedCalc to calculate the correlation coefficient of m-ELISA and flow cytometry for CD4 cell count. Altman-Bland analysis was also performed to assess the accuracy of CD4 cell quantification by the micro-a-fluidics ELISA compared to flow cytometry. The utility of micro-a-fluidic CD4 cell count for determining treatment failure or initiating antiretroviral treatment was plotted using both AIDS and immunosuppressed populations.
References
UNAIDS. UNAIDS Report on the global AIDS epidemic. http://www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/UNAIDS_Global_Report_2013_en.pdf (Data accessed: 12/2013).
Wang, S., Xu, F. & Demirci, U. Advances in developing HIV-1 viral load assays for resource-limited settings. Biotechnol. Adv. 28, 770–781 (2010).
Whitesides, G. M. The origins and the future of microfluidics. Nature. 442, 368–373 (2006).
Yager, P. et al. Microfluidic diagnostic technologies for global public health. Nature. 442, 412–418 (2006).
Tasoglu, S., Gurkan, U. A., Wang, S. & Demirci, U. Manipulating biological agents and cells in micro-scale volumes for applications in medicine. Chem. Soc. Rev. 42, 5788–5808 (2013).
Wang, S., Inci, F., De Libero, G., Singhal, A. & Demirci, U. Point-of-care assays for tuberculosis: role of nanotechnology/microfluidics. Biotechnol. Adv. 31, 438–449 (2013).
Thorsen, T., Maerkl, S. J. & Quake, S. R. Microfluidic large-scale integration. Science. 298, 580–584 (2002).
Wang, S. et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip. 11, 3411–3418 (2011).
Tasoglu, S. et al. Paramagnetic levitational assembly of hydrogels. Adv Mater. 25, 1137–1143, 1081 (2013).
Rodriguez, W. R. et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. PLoS Med. 2, e182 (2005).
Peeling, R. W., Mabey, D., Herring, A. & Hook, E. W., 3rd Why do we need quality-assured diagnostic tests for sexually transmitted infections? Nat. Rev. Microbiol. 4, 909–921 (2006).
Spacek, L. A. et al. Evaluation of a low-cost method, the Guava EasyCD4 assay, to enumerate CD4-positive lymphocyte counts in HIV-infected patients in the United States and Uganda. J. Acquir. Immune. Defic. Syndr. 41, 607–610 (2006).
Pattanapanyasat, K. et al. Evaluation of a new single-parameter volumetric flow cytometer (CyFlow(green)) for enumeration of absolute CD4(+) T lymphocytes in human immunodeficiency virus type 1-infected Thai patients. Clin. Diagn. Lab. Immunol. 12, 1416–1424 (2005).
Pattanapanyasat, K. et al. A multicenter evaluation of the PanLeucogating method and the use of generic monoclonal antibody reagents for CD4 enumeration in HIV-infected patients in Thailand. Cytometry B Clin. Cytom. 65, 29–36 (2005).
Cheng, X. et al. A microchip approach for practical label-free CD4+ T-cell counting of HIV-infected subjects in resource-poor settings. J. Acquir. Immune. Defic. Syndr. 45, 257–261 (2007).
Moon, S. et al. Integrating microfluidics and lensless imaging for point-of-care testing. Biosens. Bioelectron. 24, 3208–3214 (2009).
Gurkan, U. A. et al. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol. J. 6, 138–149 (2011).
Moon, S. et al. Enumeration of CD4(+) T-Cells Using a Portable Microchip Count Platform in Tanzanian HIV-Infected Patients. Plos One. 6, e21409 (2011).
Alyassin, M. A. et al. Rapid automated cell quantification on HIV microfluidic devices. Lab Chip. 9, 3364–3369 (2009).
World Halth Organization. Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. http://apps.who.int/iris/bitstream/10665/85321/1/9789241505727_eng.pdf (Data accessed: 12/2013).
Rizvi, I. et al. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc Natl Acad Sci U S A. 110, E1974–1983 (2013).
Tasoglu, S. et al. Exhaustion of racing sperm in nature-mimicking microfluidic channels during sorting. Small. 9, 3374–3384 (2013).
Acknowledgements
We would like to acknowledge the W.H. Coulter Foundation Young Investigator Award, RO1A1081534, RO1AI093282, U54EB15408 and R21AI087107. This work was supported by (i) the Center for Integration of Medicine and Innovative Technology (CIMIT) under U.S. Army Medical Research Acquisition Activity Cooperative Agreements DAMD17-02-2-0006, W81XWH-07-2-0011 and W81XWH-09-2-0001. We would also thank the Brigham and Women's Hospital (BWH) Biomedical Research Institute Translatable Technologies & Care Innovation Award and the Brigham Research Institute for the Bright Futures Award. Further the work was made possible by a research grant that was awarded and administered by the U.S. Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine & Advanced Technology Research Center (TATRC), at Fort Detrick, MD.
Author information
Authors and Affiliations
Contributions
S.W., D.R.K. and U.D. conceived the approach. S.W. and M.C. built the instruments. S.W., P.Z.C., M.C., U.A.G. and F.F.G. performed the experiments. R.A. wrote the mobile and desktop applications. S.T. and C.I.O. performed the modeling. All authors were involved in designing experiments, reviewing and discussing data and commenting on the manuscript.
Ethics declarations
Competing interests
U.D. is a founder of and has an equity interest in, DxNow, a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions. U.D.'s interests were reviewed and are managed by the Brigham and Women's Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Electronic supplementary material
Supplementary Information
NEW Supplementary Information File
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wang, S., Tasoglu, S., Chen, P. et al. Micro-a-fluidics ELISA for Rapid CD4 Cell Count at the Point-of-Care. Sci Rep 4, 3796 (2014). https://doi.org/10.1038/srep03796
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03796
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.