Abstract
To improve our understanding of the biological relationships among different types of cancer, we have characterized variation in gene expression patterns in a set of 1,707 samples representing 6 human cancer types (breast, ovarian, brain, colorectal, lung adenocarcinoma and squamous cell lung cancer). In the unified dataset, breast tumors of the Basal-like subtype were found to represent a unique molecular entity as any other cancer type, including the rest of breast tumors, while showing striking similarities with squamous cell lung cancers. Moreover, gene signatures tracking various cancer- and stromal-related biological processes such as proliferation, hypoxia and immune activation were found expressed similarly in different proportions of tumors across the various cancer types. These data suggest that clinical trials focusing on tumors with common profiles and/or biomarker expression rather than their tissue of origin are warranted with a special focus on Basal-like breast cancer and squamous cell lung carcinoma.
Similar content being viewed by others
Introduction
Classification and treatment of the majority of solid tumors is generally based on the tumor's tissue of origin and histological appearance (e.g. squamous cell lung cancer). In some cancer types, identification of single molecular alterations has been found to be very useful in the clinical setting due to its ability to predict treatment efficacy. For example, epidermal growth factor receptor (EGFR) mutations predict benefit from anti-EGFR drugs in lung adenocarcinoma1, amplification of epidermal growth factor receptor type 2 (HER2) predicts benefit from anti-HER2 drugs in breast cancer2, Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations predict lack of benefit from anti-EGFR drugs in colorectal cancer3 and BRCA1 mutations predict benefit from poly (ADP-ribose) polymerase 1 (PARP1) inhibitors in ovarian cancer4. Thus, searching for novel biomarkers, drug targets and better classification algorithms to individualize treatment of cancer patients is an area of active preclinical and clinical research.
In recent years, The Cancer Genome Atlas (TCGA) project has improved our understanding of the molecular alterations occurring in glioblastoma multiforme5,6, high grade serous ovarian cancer7, colorectal cancer8, squamous cell lung cancer9 and breast cancer10 and many other cancer types are being evaluated. In addition, these studies have revealed that particular molecular alterations such as TP53 mutations, MYC amplifications or CDKN2A deletions can occur in subsets of tumors of different cancer types. In fact, TCGA breast cancer project has observed that breast tumors of the Basal-like subtype share many genetic alterations with high-grade serous ovarian cancers, including TP53, RB1 and BRCA1 loss, CCNE1 and MYC amplifications and high expression of HIF1-a/ARNT, MYC and FOXM1 gene signatures10,11. Overall, these data suggest that particular treatment strategies could be effective in tumors with similar genetic alterations and/or gene expression profiles regardless of the tumor's tissue of origin11,12. Indeed, the observed benefit of anti-HER2 therapy in HER2-amplified breast and gastric cancers supports this hypothesis13,14.
To help better understand the relationships among different types of cancer, we have compared head-to-head variation in global gene expression patterns in a dataset of 1,707 samples representing 6 human cancer types.
Results
Combined microarray dataset
To study the relationships among different cancer types, we combined expression data of 17,987 genes and 1,707 samples representing 6 cancer types (glioblastoma multiforme [GBM]5,6, high-grade serous ovarian carcinoma [OVARIAN]7, lung cancer adenocarcinoma [LUAD], squamous cell lung carcinoma [SQCLC]9, colorectal adenocarcinoma [CCR]8 and breast cancer10) of the TCGA project (Fig. 1A). The cancer type with the highest gene expression variability was ovarian cancer with 9.1% of the genes showing an interquartile range of expression above 3-fold, followed closely by breast cancer (8.9%), LUAD (8.8%) and SQCLC (8.3%). CCR (4.6%) and GBM (4.5%) showed the lowest gene expression variability, suggesting that these two cancer types are biologically more homogenous.
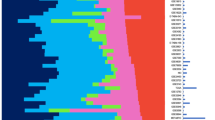
Combined gene expression microarray-based dataset of 1,707 samples representing 6 different cancer types from The Cancer Genome Atlas Project (TCGA; http://cancergenome.nih.gov/).
(A) Microarray samples analyzed from each cancer type (number of samples and color identification). (B) Principal component 1 and 2 (PC1 and PC2) loading plot using the 3,486 most variable genes. Samples have been colored based on their cancer type, except for Basal-like breast tumors (n = 98) that are colored in red. Weights of each gene for each PC can be found in Supplemental Data . (C) Correlations between PC1 or PC2 scores and expression of selected genes in the entire dataset. (D) Consensus average linkage clustering matrix for k = 3 to k = 6 of all samples and the 3,486 most variables genes. The colored bar above the matrix identifies the various cancer types represented in each k group. A single cancer type is shown in the bar if >98% of the samples of each k group are from that particular cancer type. Orange, GBM; Dark blue, OVARIAN; Light blue, CCR; Grey, SQCLC; Green, BREAST; Violet, LUAD; Red, Basal-like breast cancer.
Global gene expression landscape
To assess the global landscape of expression in the unified dataset, we performed principal components analysis (PCA)15. Brain, colorectal and breast cancer explained most of the gene expression variation displayed by the Principal Components 1 and 2 (PC1 and PC2) with samples of ovarian cancer, LUAD and SQCLC showing various levels of intermediate PC1 and PC2 scores (Fig. 1B). Strikingly, a subgroup of breast cancers almost entirely composed of the Basal-like subtype (in red color), as determined by the PAM50 subtype predictor, showed significant higher PC2 scores than the rest of breast tumors (i.e. Luminal/HER2-enriched/Normal-like) and were found close to ovarian cancers, SQCLCs and LUADs (Fig. 1B). Similar PC1 versus PC2 results were obtained from an independent gene expression-based microarray dataset of 153 samples representing breast cancer, LUAD, SQCLC and CRC (Suppl. Fig. 1).
To better understand the biological significance of PC1 and PC2, we evaluated the top-300 genes having the largest positive and negative weights for both PCs (Fig. 1C and Supplemental Data ). Gene weights are indicative of the relative contribution of each gene to the principal components. For PC1, the top-300 genes having the largest positive weight were found enriched for neuron differentiation (e.g. neuronal cell adhesion molecule [NRCAM] and N-cadherin [CDH2]), gliogenesis (e.g. SRY [sex determining region Y]-box 11 [SOX11]), cell-cell signaling (e.g. synaptotagmin IV [SYT4]) and synaptogenesis (e.g. neurexin 1 [NRXN1]), whereas the top-300 genes having the largest negative weight were found enriched for tight junctions (e.g. claudin-3 [CLDN3]), epithelial cell differentiation (i.e. FOXA1) and extracellular matrix (e.g. collagen, type XII, alpha 1 [COL12A1]). For PC2, the top-300 genes having the largest positive weight were found enriched for serine proteases (e.g. kallikrein-related peptidase 7 [KLK7]), drug metabolism (i.e. CYP3A7) and chemokines (e.g. interleukin-8 [IL8]), whereas the top-300 genes having the largest negative weight were found enriched for response to hormone stimulus (e.g. estrogen receptor [ESR1] and GATA3), cell adhesion (e.g. claudin-8 [CLDN8]) and extracellular matrix (e.g. fibronectin 1 [FN1]). Similar biological findings were obtained when the top-100, top-200 and top-400 genes were evaluated (data not shown).
Testing the molecular uniqueness of Basal-like breast cancer
The previous results suggested that Basal-like breast cancer is molecularly distinct from the other cancer types, including the rest of breast tumors. To test the level of uniqueness of Basal-like breast tumors, we performed consensus average linkage hierarchical clustering of all samples (n = 1,707) and the 3,486 most variable genes (Fig. 1D). The consensus clustering method provides quantitative and visual stability evidence for estimating the number of unsupervised classes in a dataset16. The results showed that clustering stability increased for k = 2 to k = 7 (Suppl. Fig. 2). Strikingly, Basal-like breast cancer was identified as an unsupervised class at k = 5 before colorectal cancer was separated from both lung cancer types (i.e. at k = 6) and before both lung cancer types were separated from each other (i.e. at k = 7). Overall, this result suggests that Basal-like breast cancer is a reproducible and robust cancer type.
Expression of gene signatures corresponding to human DNA regions
Somatic copy number aberrations (CNAs) in breast cancer are associated with expression in ~40% of genes17. To estimate the status of CNAs in our combined dataset, we evaluated the expression of 326 gene sets corresponding to each human chromosome and each cytogenetic band with at least one gene. These gene lists were obtained from the C1-positional gene sets of the Molecular Signature Database (Broad Institute; http://www.broadinstitute.org/gsea/msigdb/) and are helpful in identifying effects related to chromosomal deletions or amplifications.
Unsupervised hierarchical clustering of the 326 signature scores and the 1,707 samples revealed significant changes in the expression of genes located in specific DNA regions known to be aberrant in these cancer types (Fig. 2A–B). For example, high expression of arm 1q-related genes in breast cancer (including Basal-like tumors)10, or high expression of arm 13q-related genes in CCRs8, or low expression of arm 10q-related genes in GBMs5,6. In addition, we identified high expression of arm 10p-related genes and low expression of arm 5q-related genes in Basal-like breast tumors concordant with the known CNA status of these two chromosomal regions in Basal-like disease10. Finally, Basal-like breast cancers, SQCLCs and OVARIAN carcinomas clustered together consistent with the hypothesis that these three cancer types share a similar genetic profile with a special focus on 3q21-28 (amplified) and 5q13-22 (deleted) chromosomal regions (Fig. 2B).
Expression of 326 gene signatures corresponding to human DNA regions across 7 cancer types.
Signatures have been obtained from the Molecular Signatures Database (MSigDB) from the Broad Institute online website (http://www.broadinstitute.org/gsea/msigdb/collections.jsp; C1: positional gene sets). (A) Unsupervised clustering of 326 signatures scores across 1,707 samples. Each colored square on the heatmap represents the relative median signature score for each sample with highest expression being red, lowest expression being green and average expression being black. Below the array tree, samples have been colored based on their cancer type. (B) The top-10 up-regulated and down-regulated significant signatures for each cancer type (or group) are shown. These signatures were identified by performing an unpaired two-class SAM analysis between each cancer type versus the rest using the 326 signatures and a FDR = 0%.
Gene expression relationships among cancer types
To address the relationships among the 7 cancer types (i.e. Basal-like breast cancer [identified by the PAM50 subtype predictor], non-Basal-like breast cancer, CCR, GBM, SQCLC, LUAD and OVARIAN), we first identified gene expression-based centroids, representing the 7 groups, using all available genes (n = 17,987). Second, we evaluated the relationships among the different centroids within all samples (Fig. 3A), Basal-like breast tumors (Fig. 3B), OVARIAN tumors (Fig. 3C), SQCLCs (Fig. 3D), LUADs (Fig. 3E), CCRs (Fig. 3F), GBMs (Fig. 3G) and non-Basal-like breast tumors (Fig. 3H).
Transcriptomic relationships among cancer types.
Relationships have been determined by calculating the Euclidean distances of each sample to each of the 7 centroids, which represent each cancer type, using all genes of the unified dataset. Clustering has been performed after median centering the Euclidean distances of each sample. The following genomic relationships among cancer types are shown based on the following subsets of patients: (A) all patients (ALL); (B) basal-like breast cancer (BASAL-LIKE); (C) ovarian cancer (OVARIAN); (D) squamous cell lung cancer (SQCLC); (E) lung adenocarcinoma (LUAD); (F) colorectal adenocarcinoma (CCR); (G) glioblastoma multiforme (GBM); (H) non-Basal-like breast cancer (BREAST).
Strikingly, the Basal-like breast tumor centroid was found more similar to the SQCLC centroid than to the centroid of non-Basal-like breast cancer (Fig. 3B). Concordant with this, 55% of Basal-like breast tumors were found more similar (i.e. lower distances) to SQCLCs than to non-Basal-like breast cancers. When compared to the different intrinsic subtypes of breast cancer, 76%, 72% and 17% of Basal-like breast tumors were found more similar to SQCLC than to Luminal A, Luminal B and HER2-enriched breast tumors, respectively. Interestingly, Basal-like breast tumors were found more similar to both lung cancer types and to non-Basal-like breast cancers than to OVARIAN tumors (Fig. 3B).
To determine the biological processes in common between Basal-like breast cancers and SQCLC, we identified genes whose expression is found significantly expressed in both cancer types compared to luminal cancers (Luminal A and B tumors combined). Among the top 300 up-regulated genes (False Discover Rate = 0%) in Basal-like breast cancer and SQCLC, we identified genes involved in ectodermal differentiation (e.g. keratin 5, 14 and 17), inflammatory response (i.e. chemokine [C-X-C motif] ligand 1 [CXCL1] and CXCL3) and cell cycle (e.g. cyclin E1 [CCNE1] and centromere protein A [CENPA]). Among the top 300 down-regulated genes, we identified genes involved in the response to hormone stimulus (e.g. estrogen receptor [ESR1] and GATA3), mammary gland development (e.g. prolactin receptor [PRLR] and ERBB4) and microtubule-based process (e.g. kinesin family member 12 [KIF12] and microtubule-associated protein tau [MAPT]). This data is concordant with the histological appearance and the immunohistochemical expression of ER, keratins 5/6 and the proliferation-related biomarker Ki67 in a Basal-like breast tumor, a SQCLC with a Basal-like profile and a breast Luminal A tumor (Fig. 4).
Immunohistochemical (IHC) and PAM50 gene expression analyses of a Basal-like breast cancer, a SQCLC with a Basal-like profile and a Luminal A breast cancer.
Hematoxylin/eosine (H/E); Estrogen receptor (ER) expression; Keratin 5/6 (KRT5/6) expression; Proliferation-related Ki-67 expression. Each colored square on the heatmap below the IHC images represents the relative transcript abundance (in log2 space) of each PAM50 gene with highest expression being red, lowest expression being green and average expression being black.
Multiclass tumor prediction
To identify genes that are distinctive of each cancer type, including Basal-like breast cancer, we performed ClaNC, a nearest centroid-based classifier that balances the number of genes per class (Fig. 5A). A 126-gene signature (18 genes per cancer type) was established from the smallest gene set with the lowest cross validation and prediction error (2.0%) (Fig. 5B). Among the various cancer types, Basal-like breast cancers and SQCLCs showed the highest prediction error (7.1% and 15.6%) and the majority of misclassified SQCLCs (n = 5, 71.4%) were identified as Basal-like breast cancer. Of note, two previously identified diagnostic biomarkers of serous ovarian cancer (Wilm's tumor [WT]-1)18 and lung adenocarcinoma (thyroid nuclear factor 1 [TITF-1])19 were found in the 18-gene list of these two cancer types (Fig. 5C).
Identification of a cancer type-specific 126-gene signature.
(A) Clustering of the 126-gene scores obtained after Classification by Nearest Centroids (ClaNC) analysis. Discriminative genes are shown in red and in green color. (B) Training error of the 126-gene signature across the 7 cancer-types. (C) Genes whose high (red) or low (green) expression define each cancer type. Gene Symbols are shown first, followed by the Entrez GeneIDs.
Common patterns of gene expression across cancer types
Although each cancer type is molecularly distinct, we sought to identify groups of genes (i.e. gene signatures) with independent patterns of variation. To accomplish this, we clustered all samples with the 3,486 most variable genes (Fig. 6) and identified 19 gene clusters of at least 10 genes and an intraclass correlation coefficient >0.70 ( Supplemental Data ). Among them, we identified gene signatures tracking lymphocyte activation/infiltration (e.g. CD8A and CD2), ectodermal development (e.g. keratin 6B and 15), interleukin-8 pathway (e.g. IL8 and CXCL1), tight junctions (e.g. claudin-3 and occludin), proliferation (e.g. budding uninhibited by benzimidazoles 1 homolog [BUB1] and CENPA) and interferon-response pathways (e.g. STAT1 and interferon-induced protein with tetratricopeptide repeats 1 [IFIT1]) (Fig. 6).
Unsupervised hierarchical clustering of 7 different cancer types and the 3,486 most variable genes.
(A) Tumor samples have been ordered based on the consensus clustering matrix (Fig. 1D). Each colored square on the heatmap represents the relative median transcript abundance (in log2 space) for each sample with highest expression being red, lowest expression being green and average expression being black. (B) Expression of selected gene clusters tracking several cancer-related biological processes. Clusters were selected based on the following criteria: at least 10 genes and an intraclass correlation coefficient >0.70. Colored bars to the right identify the locations of the selected clusters. The gene list of the 19 gene clusters identified can be found in Supplemental Data . Gene symbols and Entrez GeneIDs are shown on the right.
Common patterns of gene signature expression across cancer types
Similar to the previous analysis, we determined the expression scores of 329 gene signatures (or modules)20 in all samples, including 115 previously published signatures and then performed an unsupervised hierarchical clustering (Fig. 7). Thirteen clusters of at least 5 signatures and an intraclass correlation coefficient >0.70 were identified. These groups of gene signatures were found to track various types of biological processes/features likely coming from the tumor cell, the microenvironment or both. Interestingly, the expression of signatures tracking microenvironment-related (e.g. lymphocyte activation/infiltration) biological processes were found to be less cancer type specific than the expression of gene signatures tracking tumor-related biological processes (e.g. proliferation).
Unsupervised hierarchical clustering of the 7 different cancer types and 329 gene signature scores.
(A) 329 gene signatures and all tumor samples. Each colored square on the heatmap represents the relative signature score (in log2 space) for each sample with highest expression being red, lowest expression being green and average expression being black. On the right, selected signature groups were selected. Black bars to the right identify the approximate locations of the selected groups. The signature scores of each sample can be found in Supplemental Data .
To illustrate the overlap among cancer types regarding the expression of a single signature, we evaluated 6 previously identified gene signatures that are known to track various cancer-related and stromal/microenviroment-related biological processes related to breast cancer biology21,47,48,49,50,51. The results showed that high expression of these signatures (i.e. the top 20% expressers in the unified dataset) occurs across all cancer types, albeit with different proportions (Fig. 8). Of note, the TP53 signature21, which was trained in a previously reported breast cancer dataset, predicted TP53 somatic mutations in the combined TCGA dataset (area under the receiver operating characteristic curve = 0.782; Suppl. Fig. 3). Moreover, the scores of the previously reported PTEN-loss signature were found correlated with INPP4B (correlation coefficient = −0.424, p-value < 0.0001) and phospho-4E-BP1 (correlation coefficient = 0.368, p-value < 0.0001) protein expression in the TCGA breast cancer dataset (Suppl. Fig. 4).
Expression of 6 selected gene signatures (SIGN) representing various cancer-related biological processes in the unified dataset.
For each gene signature, samples have been rank ordered based on their gene signature score (red, black and green represent above, equal and below the median score). Samples from each cancer type are shown below the gene signature score heatmap. Percentages of samples present in the top 20% expressers in the entire dataset are shown on the left. PROLIF, proliferation; EMT, epithelial to mesenchymal transition; TP53MUT, P53 mutation.
Breast cancer intrinsic subtyping of non-breast tumors
To evaluate if the breast cancer ‘intrinsic’ profiles (Luminal A, Luminal B, HER2-enriched and Basal-like) can be identified in non-breast tumors, we performed breast cancer intrinsic subtyping of non-breast cancer types using the PAM50 and Claudin-low subtype predictors22,23. Interestingly, all the breast cancer ‘intrinsic’ profiles were identified albeit with different proportions (Table 1). For example, the Basal-like profile was identified in 55% and 53% of SQCLC and ovarian cancers, respectively, whereas virtually all colorectal cancers (99%) and most lung adenocarcinomas (59%) showed the HER2-enriched profile. Of note, 28% of ovarian cancers and 24% of SQCLC tumors also showed the HER2-enriched profile. Finally, the Claudin-low profile was identified in 20% and 16% of SQCLCs and LUADs, respectively.
To provide further evidence, we performed breast cancer intrinsic subtyping of non-breast cancer types in two independent datasets (Suppl. Fig. 5 and 6). First, we evaluated a publicly available microarray dataset (GSE23768) that includes 153 samples of breast cancer (n = 84), CCR (n = 26), SQCLC (n = 22) and LUAD (n = 21)24 (Suppl. Fig. 5). Similar to previous results, 92.3% and 7.7% of all CCRs were identified as HER2-enriched and Basal-like and 59.1% and 27.3% of SQCLCs were identified as Basal-like and Claudin-low, respectively.
Finally, we evaluated PAM50 gene expression in a dataset from Vall d'Hebron Institute of Oncology that includes 125 formalin-fixed paraffin-embedded tumor samples of breast cancer (n = 53) and CCR (n = 72) (Suppl. Fig. 6). Similar to previous results, 88.9% and 11.1% of all CCRs were identified as HER2-enriched and Basal-like, respectively. Of note, ERBB2 was not found overexpressed in these CRCs HER2-enriched tumors, except for 1 case (1.4%), which we found highly amplified in HER2 by FISH (ratio gene/chromosome = 7.23) (Suppl. Fig. 7).
Discussion
In this study, we have evaluated global gene expression data of 6 human cancer types from the TCGA project and made the following observations. First, a group of breast cancers, virtually all of the Basal-like subtype, have a unique and distinct profile as the profile of any of the other cancer types, including the rest of breast tumors. Second, although Basal-like breast tumors and high grade serous ovarian carcinomas may share similar genetic alterations10, our transcriptomic analyses show that the majority of Basal-like tumors are more similar to SQCLCs than to high grade serous ovarian carcinomas and the rest of breast tumors. Indeed, when compared with each breast cancer intrinsic subtype, ~70% Basal-like tumors were found more similar to SQCLC than Luminal A and B breast tumors. Third, we identified cancer type specific genes and confirmed the association of WT-1 and TITF-1 expression with the identification of serous ovarian cancer and LUAD, respectively. Finally, we provided evidence that distinct gene signatures can be found expressed regardless of the cancer type. For example, we identified the Basal-like breast cancer profile in the majority of SQCLC and ovarian cancers, whereas the HER2-enriched breast cancer profile was found expressed in a subset of ovarian cancers and SQCLCs and the majority of LUADs and CCRs.
The observation that Basal-like breast cancer is remarkably different from the rest of breast tumors argues for two distinct cells of origin in the mammary gland25. In fact, ducts and lobules in the normal human breast are lined by two cell layers, an inner/luminal cell population that expresses luminal keratins such as keratins 8, 18 and 19 as well as BCL2, MUC1 and estrogen receptor and a distinct outer cell layer, named the basal layer, which is composed of cells that exhibit features of both epithelial and smooth muscle cells such as expression of keratins 5, 14 and 17, smooth muscle actin and p63. However, basal keratins are also expressed in a subpopulation of luminal cells26,27 and preclinical mouse work has shown conflicting data regarding the cell of origin of Basal-like breast tumors28,29,30,31 (i.e. luminal progenitor versus bipotent progenitor/stem cell). In any case, our observation that the majority of Basal-like tumors are more similar to SQCLC than to the rest of breast tumors, especially to the luminal tumors, suggests that the cell of origin of Basal-like breast tumors has similar features and location, in an epithelial layer, as the cell of origin of SQCLCs, which is the basal cell present in the airway basal lamina32.
The transcriptomic similarities observed here between Basal-like breast cancers and SQCLCs are also found at the DNA level. Indeed, the most frequently mutated genes in Basal-like breast tumors (i.e. TP53, PIK3CA and RB1) are also found in the top 10 significantly mutated genes in SQCLC. Of note, the most frequently mutated gene in both tumors types is TP53 with a prevalence of ~80%. In addition, both cancer types also share many focal regions of DNA amplification or deletion ( Supplemental Data ). Among 28 significant focal DNA regions of amplification in Basal-like breast cancer identified in the TCGA breast cancer project10, 7 (25%) are also found in SQCLC: 8q24.21 (e.g. MYC), 8p11.23 (e.g. FGFR1), 7p11.2 (e.g. EGFR), 12p13.33 (e.g. FOXM1), 15q26.3 (e.g. ALDH1A3), 21q21.1 (e.g. miRNA-let-7c), 11q13.3 (e.g. FGF3 and FGF4). Among 36 significant focal DNA regions of deletion in Basal-like breast cancer, 13 (36%) are also found in SQCLC such as 10q23.31 (e.g. PTEN), 10q26.3 (e.g. INPP5A), 11q25 (e.g. ATM), 4p16.3 (e.g. INPP4B), 9p21.3 (e.g. CDKN2A and CDKN2B) and 5q11.2 (e.g. RAD17). Finally, at the clinical level, both cancer types have a poor prognosis with a high propensity for metastatic spread33,34,35. In terms of treatment response, both show high sensitivity to platinum-based therapies36,37 although resistant disease easily emerges38.
The TGCA breast cancer project has previously found common genetic alterations between Basal-like breast cancer and high-grade serous ovarian carcinomas10,11. Concordant with this, we observed that most ovarian cancers show a Basal-like profile and are more similar to Basal-like breast cancer than to the other cancer types evaluated; conversely, Basal-like breast tumors are more similar to both lung cancers and non-Basal-like breast cancer than to ovarian cancer. The potential explanation for this non-reciprocity despite that serous ovarian cancers and Basal-like breast tumors share many alterations at the DNA level is currently unknown but might well be related to the fact that the cell of origins of these two tumor types might have different biological functions and/or epigenetic programs in its respective organ. Although targeting a similar genetic alteration in two different cancer types might be a successful strategy in some cases (i.e. HER2 amplification in HER2+ gastric and breast cancers)12,13, tumor cell context might also be important in other cases as highlighted by the limited response observed so far to BRAF inhibitors of BRAF (V600E) mutated colorectal cancers compared to BRAF-mutated melanomas39.
Despite the differences in global gene expression profiles, ovarian cancer does have a similar prevalence of TP53 mutations and RB1 as Basal-like breast cancer and SQCLC. In addition, the three cancer types also share 3 focal regions of DNA amplification (8q24.21, 12p13.33 and 15q26.3) and 10 focal regions of DNA deletion (2q22.1, 2q37.3, 5q11.2, 9p21.3, 10q23.31, 10q26.3, 11q25, 16p13.3, 18q23 and 19p13.3) ( Supplemental Data ). Interestingly, our expression analysis of DNA chromosomal regions and cytogenic bands has also identified the 5q13-22 region as a commonly deleted across the three cancer types. Interestingly, this DNA region contains BRCA1-dependent DNA repair genes such as RAD17, RAD50 and RAP80 and its deletion has been shown to increase sensitivity to a PARP inhibitor and carboplatin in preclinical models of Basal-like breast cancer40.
Over the years, the intrinsic subtypes of breast cancer (Luminal A, Luminal B, HER2-enriched and Basal-like) have shown significant differences in terms of survival, response to therapies and dissemination patterns33. Identification of these ‘intrinsic’ profiles in non-breast cancer types revealed interesting associations. For example, virtually all CRCs in the TCGA and in two independent datasets were identified as HER2-enriched despite ERBB2 not being overexpressed or amplified8,41. This result suggests that another oncogene or driver might be inducing a similar activation of the downstream signaling pathways, including MAPK/RAS pathways, in a similar manner as ERBB2. Interestingly, we have previously shown that the HER2-enriched subtype can be identified in HER2-nonamplified breast cancers (especially within the so-called triple-negative disease) and these tumors tend to show high expression of EGFR42. Another interesting observation has been the identification of the Luminal A and B breast cancer ‘intrinsic’ profiles in 11% of lung adenocarcinomas, suggesting that these tumors are enriched for estrogen receptor-regulated biological processes. Interestingly, substantial preclinical evidence supports a role for hormonal influence on lung cancer43,44 and treatment with estrogen plus progestin in women has shown to increase deaths from lung cancer45.
Although we could identify common pattern of gene (or signature) expression across different cancer types, each cancer type (including Basal-like breast cancer) is a unique molecular entity when global gene expression analyses are performed. This prompted us to identify a robust 126-gene signature (18 genes per cancer type) that can identify the 6 cancer types. Interestingly, the list included known cancer type specific biomarkers such WT-1 (for serous ovarian cancer) and TITF-1 (for lung adenocarcinoma). Other biomarkers somewhat related to specific cancer types were keratin 20 (KRT20) for CRCs, prolactin receptor (PRLR) for non-Basal-like breast cancers, forkhead box C1 (FOXC1) for Basal-like breast cancers, tumor protein p63 (TP63, also known as TP73L) for SQCLCs, various surfactant associated proteins (SFTPA2, SFTPC and SFTPD) for LUADs and oligodendrocyte transcription factor 2 (OLIG2) for GBMs.
To conclude, molecular screening for specific alterations in the tumor cells and/or the tumor microenviroment such as DNA mutations, DNA amplifications and RNA and protein expression, is becoming commonplace in many parts of the world46. Patients whose tumor has a specific biological alteration that might be inhibited by a novel biological drug are being incorporated in early phase clinical trials regardless of the cancer type. Indeed, a subset of gastric and breast cancers showing amplification of HER2 benefit from anti-HER2 therapies13,14. At the same time, various cancer types benefit from anti-angiogenic (i.e. bevacizumab) and immune enhancers (PD-L1 antibody)47,48. Although this strategy shows promise as evidence by the results of some studies, it is still unknown how best to approach it. Our data argues in favor of targeting common molecular alterations of the tumor and/or the microenvironment regardless of the tumor's tissue of origin with a special focus on Basal-like breast and squamous cell lung cancer.
Methods
Gene expression data from TCGA
All gene expression microarray data is publicly available and can be obtained from the TCGA portal (https://tcga-data.nci.nih.gov/tcga/). We evaluated a total of 1,580 samples from published cancer types from TCGA including glioblastoma multiforme5,6 (GBM, n = 202), high-grade serous ovarian carcinoma7 (named here ovarian cancer, n = 489), breast cancer10 (n = 547), colorectal cancer8 (CCR, n = 220) and squamous cell lung carcinoma9 (SQCLC, n = 122). In addition, we also included 127 unpublished samples from the TCGA lung adenocarcinoma (LUAD) project that are already publicly available in the TCGA portal for a total of 1,707 samples. Of note, all microarrays were performed at the University of North Carolina (UNC) under the same protocol and platform (Agilent Technology, Santa Clara, CA). Briefly, 1 to 2 ug of total RNA sample and Stratagene Universal Human Reference were amplified and labeled using Agilent's Low RNA Input Linear Amplification Kit. The total yield of amplified RNA (aRNA) and Cy dye incorporation was measured by NanoDrop. Sample and reference 7–10 ug of each) were co-hybridized to a Custom Agilent 244K Gene Expression Microarray. Arrays were scanned on an Agilent Scanner and probe information was obtained with Agilent's Feature Extraction Software.
All microarray data was downloaded from the UNC UMD website (https://genome.unc.edu/). The probes or genes for all analyses were filtered by requiring the lowess normalized intensity values in both sample and control to be >10. The normalized log2 ratios (Cy5 sample/Cy3 control) of probes mapping to the same gene (Entrez ID as defined by the manufacturer) were averaged to generate independent expression estimates of a total of 17,987 genes. No significant batch effects based on the microarray chip barcode were observed (Suppl. Fig. 8–9).
Gene expression signatures
In the unified dataset, we evaluated 329 gene signatures using the gene lists and algorithms previously described20,21,23,49,50,51. In addition, we evaluated the percentage of tumors within a cancer type that falls within the top-20% expressers in the unified dataset for the following gene sigantures: a TP53 mutation signature21, a VEGF13-hypoxia signature49, an immune cell signature19, a 11-gene proliferation signature23, a PTEN loss signature50 and an epithelial-to-mesenchymal transition (EMT) signature51.
To identify the breast cancer ‘intrinsic’ subtype in non-breast tumors (Luminal A, Luminal B, HER2-enriched, Basal-like, Claudin-low and Normal-like), we used the PAM50 intrinsic subtype classifier23 and the Claudin-low subtype predictor22 after gene centering the combined dataset onto the breast cancer dataset. To ensure subtyping of non-breast cancers was correctly implemented, we compared the subtype calls obtained in breast cancers compared to the subtype calls provided in the TCGA breast cancer datasets.
Subtype identification
For all cancer types, except LUAD, we used the subtype calls as provided in each TCGA publication5,6,7,8,9. For LUADs, we used the tumor centroids provided in Wilkerson et al.52 and assigned a subtype call (Bronchioid, magnoid and squamoid) for each LUAD using Single Sample Predictor (SSP).
Immunohistochemical (IHC) analyses
We selected 3 tumors representing a Basal-like breast cancer, a SQCLC with a PAM50 Basal-like profile and a Luminal A breast cancer. To confirm the intrinsic subtype of the two breast tumors and the Basal-like profile of the lung cancer, we performed expression analyses of the 50 PAM50 genes (and 5 house-keeping genes) using the nCounter Nanostring platform after purifying ~100 ng of total RNA from the same tumor block where IHC analyses were performed. Immunohistochemical staining using the avidin-biotin-peroxidase technique was performed for each antibody. Five- micrometer-thick sections were cut from the tissue specimens and placed on poly-L-lysine–coated glass slides. Sections were deparaffined by xylene and rehydrated in graded alcohol. Endogenous peroxidase was blocked by immersing the sections in 0.1% hydrogen peroxidase in absolute methanol for 20 min. For antigen retrieval, the tissue sections were heated in a pressure cooker in citric acid monohydrate 10 mM, pH 6.0, for 5 min and then incubated with the primary antibody at room temperature. IHC was performed with Benchmark XT (Ventana Medical Systems, Inc, Tucson, AZ). The primary antibodies and dilutions used were: anti-ER (Ventana Medical Systems, prediluted), anti-Ki-67 (Ventana Medical Systems, prediluted), anti-CK5/6 (Biocare, prediluted). All slides were hematoxylin counterstained, dehydrated and mounted. Omitting the primary antibody performed negative controls.
Evaluation of two independent gene expression-based datasets
The first dataset is a publicly available microarray dataset (GSE23768, Affymetrix Human Exon 1.0 ST Array)24 that includes 153 samples of breast cancer (n = 84), CCR (n = 26), SQCLC (n = 22) and LUAD (n = 21). Normalized data was obtained from Gene Expression Omnibus and only the most variable probes (i.e. highest inter-quartile range) for each gene were considered for further analyses.
The second dataset is an in-house dataset of VHIO that includes 53 formalin-fixed paraffin-embedded (FFPE) breast samples representing all the subtypes and 72 primary untreated FFPE CCRs. We performed expression analyses of 49 of the 50 PAM50 genes (and 5 house-keeping genes) using the nCounter Nanostring platform after purifying ~100 ng of total RNA.
Statistical analysis
From the combined microarray dataset, we selected the most variable genes within each cancer-type (breast, ovarian, LUAD, SQCLC, GBM and CCR) by selecting 3,486 genes with an interquartile range (i.e. difference between the upper and lower quartiles), which is a measure of statistical dispersion, of at least 3-fold ( Supplemental Data ). To estimate the ‘distance’ of each sample to each cancer type centroid, we calculated the Euclidean distance using SSP. Differences between ‘distances’ of each sample to two centroids were used to determine the grade of similarity.
All microarray cluster analyses were displayed using Java Treeview version 1.1.4r2 and can be found in supplemental materials. Average-linkage hierarchical clustering was performed using Cluster v3.053. Biologic analysis of microarray data was performed with DAVID annotation tool (http://david.abcc.ncifcrf.gov/)54. Only significant (p < 0.001) Gene Ontology enrichments are shown in the text. Consensus clustering plus (http://bioc.ism.ac.jp/2.6/bioc/html/ConsensusClusterPlus.html)16 and principal complement plots of gene expression data were performed using R (http://cran.r-project.org).
Change history
11 February 2015
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has been fixed in the paper.
References
Lynch, T. J. et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. New Engl J Med 350, 2129–2139 (2004).
Baselga, J. et al. Pertuzumab plus Trastuzumab plus Docetaxel for Metastatic Breast Cancer. New Engl J Med 366, 109–119 (2012).
Tejpar, S. et al. Association of KRAS G13D Tumor Mutations With Outcome in Patients With Metastatic Colorectal Cancer Treated With First-Line Chemotherapy With or Without Cetuximab. J Clin Oncol 30, 3570–3577 (2012).
Fong, P. C. et al. Poly(ADP)-Ribose Polymerase Inhibition: Frequent Durable Responses in BRCA Carrier Ovarian Cancer Correlating With Platinum-Free Interval. J Clin Oncol 28, 2512–2519 (2010).
Network, T. C. G. A. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Verhaak, R. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer cell 19, (2010).
Network, T. C. G. A. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Network, T. C. G. A. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Network, T. C. G. A. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
Network, T. C. G. A. Comprehensive molecular portraits of human breast tumors. Nature 490, 61–70 (2012).
Ellis, M. J. & Perou, C. M. The Genomic Landscape of Breast Cancer as a Therapeutic Roadmap. Cancer Discov 3, 27–34 (2013).
Cortes, J. et al. New Approach to Cancer Therapy Based on a Molecularly Defined Cancer Classification. CA Cancer J Clin (2013 In Press).
Bang, Y.-J. et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010).
Baselga, J. et al. Phase II Study of Efficacy, Safety and Pharmacokinetics of Trastuzumab Monotherapy Administered on a 3-Weekly Schedule. J Clin Oncol 23, 2162–2171 (2005).
Ringner, M. What is principal component analysis? Nat Biotech 26, 303–304 (2008).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Goldstein, N. S., Bassi, D. & Uzieblo, A. WT1 Is an Integral Component of an Antibody Panel to Distinguish Pancreaticobiliary and Some Ovarian Epithelial Neoplasms. Am J Clin Pathol 116, 246–252 (2001).
Anagnostou, V. K. et al. Molecular classification of nonsmall cell lung cancer using a 4-protein quantitative assay. Cancer 118, 1607–1618 (2012).
Fan, C. et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics 4, 3 (2010).
Troester, M. et al. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer 6, 276 (2006).
Prat, A. et al. Phenotypic and Molecular Characterization of the Claudin-low Intrinsic Subtype of Breast Cancer. Breast Cancer Res 12, R68 (2010).
Parker, J. S. et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol 27, 1160–1167 (2009).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Prat, A. & Perou, C. M. Mammary development meets cancer genomics. Nat Med 15, 842 (2009).
Gusterson, B., Ross, D., Heath, V. & Stein, T. Basal cytokeratins and their relationship to the cellular origin and functional classification of breast cancer. Breast Cancer Res 7, 143–148 (2005).
Rakha, E. A., Reis-Filho, J. S. & Ellis, I. O. Basal-Like Breast Cancer: A Critical Review. J Clin Oncol 26, 2568–2581 (2008).
Liu, X. et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Nat Acad Sci USA 104, 12111–12116 (2007).
Molyneux, G. et al. BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell stem cell 7, 403–417 (2010).
Chaffer, C. L. & Weinberg, R. A. Cancer Cell of Origin: Spotlight on Luminal Progenitors. Cell stem cell 7, 271–272 (2010).
Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15, 907 (2009).
Sutherland, K. D. & Berns, A. Cell of origin of lung cancer. Mol Oncol 4, 397–403 (2010).
Scaltriti, M. et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Nat Acad Sci USA 108, 3761–3766 (2011).
Harrell, J. et al. Genomic analysis identifies unique signatures predictive of brain, lung and liver relapse. Breast Cancer Res Treat 132, 523–535 (2012).
Ou, S. H. I., Zell, J. A., Ziogas, A. & Anton-Culver, H. Prognostic factors for survival of stage I nonsmall cell lung cancer patients. Cancer 110, 1532–1541 (2007).
Silver, D. P. et al. Efficacy of Neoadjuvant Cisplatin in Triple-Negative Breast Cancer. J Clin Oncol 28, 1145–1153 (2010).
Martin, M. et al. Genomic predictors of response to doxorubicin versus docetaxel in primary breast cancer. Breast Cancer Res Treat 128, 127–136 (2011).
Balko, J. et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med 18, 1052–1059 (2013).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Weigman, V. et al. Basal-like Breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy and patient survival. Breast Cancer Res Treat 133, 865–880 (2012).
Sclafani, F. et al. HER2 in high-risk rectal cancer patients treated in EXPERT-C, a randomized phase II trial of neoadjuvant capecitabine and oxaliplatin (CAPOX) and chemoradiotherapy (CRT) with or without cetuximab. Ann Oncol (2013 In Press).
Prat, A. et al. Molecular Characterization of Basal-Like and Non-Basal-Like Triple-Negative Breast Cancer. Oncologist 18, 123–133 (2013).
Stabile, L. P. et al. Combined Targeting of the Estrogen Receptor and the Epidermal Growth Factor Receptor in Non–Small Cell Lung Cancer Shows Enhanced Antiproliferative Effects. Cancer Res 65, 1459–1470 (2005).
Márquez-Garbán, D. C., Chen, H.-W., Goodglick, L., Fishbein, M. C. & Pietras, R. J. Targeting Aromatase and Estrogen Signaling in Human Non-Small Cell Lung Cancer. Ann N Y Acad Sci 1155, 194–205 (2009).
Chlebowski, R. T. et al. Oestrogen plus progestin and lung cancer in postmenopausal women (Women's Health Initiative trial): a post-hoc analysis of a randomised controlled trial. Lancet 374, 1243–1251 (2009).
Rodon, J. et al. Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol 9, 359–366 (2012).
Braghiroli, M. I., Sabbaga, J. & Hoff, P. M. Bevacizumab: overview of the literature. Expert Review of Anticancer Therapy 12, 567–580 (2012).
Brahmer, J. R. et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. New Engl J Med 366, 2455–2465 (2012).
Hu, Z. et al. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med 7, 9 (2009).
Saal, L. H. et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A 104, 7564–7569 (2007).
Taube, J. H. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A 107, 15449–15454 (2010).
Wilkerson, M. D. et al. Differential Pathogenesis of Lung Adenocarcinoma Subtypes Involving Sequence Mutations, Copy Number, Chromosomal Instability and Methylation. PLoS ONE 7, e36530 (2012).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95, 14863–14868 (1998).
Dennis, G. et al. DAVID: database for annotation, visualization and Integrated discovery. Genome Biol 4, R60 (2003).
Acknowledgements
This work was supported by funds from the ROCHE Spanish Grant for Translational Research (A.P.) and Instituto de Salud Carlos III - PI11/00917 (J.T.).
Author information
Authors and Affiliations
Contributions
A.P., B.A. and J.C. contributed to experimental design. A.P., V.P. and P.G. were responsible for performing experiments. A.P., B.A., V.P. and C.F. contributed to data analysis. A.P., B.A., C.F., V.P., M.V., P.G., S.D., A.V., J.R., H.G.P., P.N., S.R.C., J.M.D., E.F., J.T. and J.C. contributed to manuscript preparation.
Ethics declarations
Competing interests
A.P. reports an uncompensated advisory role for Nanostring Technologies.
Electronic supplementary material
Supplementary Information
TCGA GEP Combined Matrix
Supplementary Information
Supplemental Data
Supplementary Information
Suppl. Figures 1–8
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Prat, A., Adamo, B., Fan, C. et al. Genomic Analyses across Six Cancer Types Identify Basal-like Breast Cancer as a Unique Molecular Entity. Sci Rep 3, 3544 (2013). https://doi.org/10.1038/srep03544
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03544
This article is cited by
-
Discovery of primary prostate cancer biomarkers using cross cancer learning
Scientific Reports (2021)
-
RETRACTED ARTICLE: Production of viable chicken by allogeneic transplantation of primordial germ cells induced from somatic cells
Nature Communications (2021)
-
Whales, fish and Alaskan bears: interest-relative taxonomy and kind pluralism in biology
Synthese (2021)
-
Direct formalin fixation induces widespread transcriptomic effects in archival tissue samples
Scientific Reports (2020)
-
A functional role for the cancer disparity-linked genes, CRYβB2 and CRYβB2P1, in the promotion of breast cancer
Breast Cancer Research (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.