Abstract
Near Eastern wild boars possess a characteristic DNA signature. Unexpectedly, wild boars from Israel have the DNA sequences of European wild boars and domestic pigs. To understand how this anomaly evolved, we sequenced DNA from ancient and modern pigs from Israel. Pigs from Late Bronze Age (until ca. 1150 BCE) in Israel shared haplotypes of modern and ancient Near Eastern pigs. European haplotypes became dominant only during the Iron Age (ca. 900 BCE). This raises the possibility that European pigs were brought to the region by the Sea Peoples who migrated to the Levant at that time. Then, a complete genetic turnover took place, most likely because of repeated admixture between local and introduced European domestic pigs that went feral. Severe population bottlenecks likely accelerated this process. Introductions by humans have strongly affected the phylogeography of wild animals, and interpretations of phylogeography based on modern DNA alone should be taken with caution.
Similar content being viewed by others
Introduction
Phylogeographic studies use phylogenetic methods and geographical contexts to reveal evolutionary histories of species to follow migrations and contraction/expansion processes1,2. Recent studies of mitochondrial DNA (mtDNA) of wild boar (Sus scrofa) found strong phylogeographic structure across its entire distribution range with European, Near Eastern and East Asian pigs falling into separate clades3,4,5,6,7. This structure has enabled researchers to infer past events in the history of wild boars, such as postglacial recolonisation of northern Europe from southern refugia8. Violation of this phylogeographic structure was used to infer human movements and translocations across Europe and the Pacific4,5,9.
During the Epipaleolithic and Neolithic periods, humans introduced wild boars into various regions across Eurasia (e.g.10,11,12). Since domestication, however, the assumption is that humans have mainly translocated domestic pigs rather than wild boars. It was remarkable, then, that Giuffra et al.3 reported that three modern wild boars from Israel possessed a European mtDNA haplotype. In contrast, studies have shown that modern wild boars from other Near Eastern countries (i.e., Egypt, Syria, Turkey, Armenia, Iraq and Iran) possess Near Eastern haplotypes5,7,9.
Patterns observed using only modern mtDNA might not accurately represent earlier phylogeographic events such as migrations, translocations and extinctions (e.g.13,14,15). Therefore, ancient DNA is a powerful tool for revealing population histories, and can shed light on past events (e.g.16,17,18). Through the use of ancient DNA, we aim to elucidate the apparent paradox of the modern Israeli wild boar, and to determine just how far back this inconsistency extends.
Today, wild boars are found in the Mediterranean region of Israel and south of the Dead Sea19. They have been part of the fauna of the southern Levant since at least the middle Pleistocene, ca.0.78 Mya20. Pigs were domesticated in the Neolithic21,22 and their remains are found at most archaeological sites in Israel. Their frequency, however, varies greatly between sites as well as between periods (e.g.23,24). Variation in the number of pig bones, and therefore in the past consumption of pork, depends on several factors, including climate, modes of life, as well as cultural and religious preferences25,26,27. Pig remains, however, are especially abundant in the Israeli archaeological record in four periods: Middle Bronze II (ca. 2000 BCE–1550 BCE); Iron I (ca. 1150-800 BCE) faunal assemblages in Philistia (the southern coastal plain) (e.g.23,28,29,30); assemblages from non-Jewish sites from the Roman (634-324 BCE) and Byzantine periods (324-63 BCE)23,27. There is extensive archaeological evidence showing that during all these periods there were contacts between the Levant and Europe (e.g.31,32,33; needless to say, connections became stronger with time).
In order to resolve the phylogeographic inconsistency that the modern Israeli wild boars have a different mtDNA signature from their surroundings, we examined a combination of modern and ancient DNA sequences of suids from Israel. We analysed DNA from a series of modern wild boars from museum collections representing their full biogeographical distribution across the country, in order to assess the observation made by Giuffra et al.3 and examine whether their results reflected a genetic anomaly or a widespread pattern. Ancient DNA data enabled us to test whether this was a case of shifting spatial and temporal patterns of haplotypes, and if so to pinpoint when it had started.
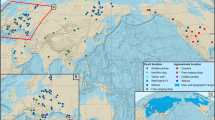
For the modern wild boar samples, we amplified a combination of mtDNA control region (CR), and nuclear Melanocortin receptor 1 (MC1R) gene. This is the first time that Near Eastern wild boars have been sequenced for the MC1R gene that is associated with coat colour variation (e.g.34,35). We chose this nuclear marker because a large geographical dataset is available for this gene. Moreover, recent studies showed that pig coat colour is the result of strong selective pressures (e.g.36,37). Thus European and Asian pigs can be distinguished according to their MC1R gene. Using ancient DNA techniques, we studied domestic pig bones from a diachronic series of Israeli archaeological assemblages dating from the Neolithic period to medieval times (Fig. 1). For the ancient samples we analysed a short fragment of the mtDNA CR. We then compared our results to phylogenetic data on Asian and European pigs to elucidate the biogeographic affinities of our samples.
Results
Modern DNA
We analysed the DNA from 25 museum specimens of modern wild boars from Israel using mtDNA CR (see Supplementary Table S1). The phylogenetic tree of mtDNA CR was based on 621 bp, and is in agreement with previous studies of Sus scrofa5,6,7,9. Three supported clades are known – one European (with Italian samples forming a divergent clade at the base of the group), and two Near Eastern – while the rest of the tree consists of East Asian pigs (Fig. 2). All 25 modern wild boars from Israel as well as one Israeli boar sequence from GenBank (accession number AF136558) (Fig. 2) fell within the European clade. Very little genetic diversity was found in the mtDNA CR between these wild boar sequences, with 21 samples sharing the same haplotype.
A Bayesian phylogenetic tree based on 621 bp of the mitochondrial CR with 100 haplotypes, representing 197 modern pig sequences (domestic and wild boars). Desert Warthog (Phacochoerus aethiopicus) serves as an outgroup. Bayesian posterior probabilities (above 0.8), and bootstrap supports (above 50%) are shown below and above the branches respectively. The haplotype names are after extraction numbers (MMP for samples extracted in this study). N# indicates how many samples have the same sequence. For more details see Supplementary Table S1.
We also analysed the nuclear MC1R region in 17 modern wild boar specimens from Israel. A median-joining network, based on 963 bp of MC1R region demonstrates the differences between European and Asian suids, and confirms that extant wild boars from Israel possess a European wild boar haplotype (identical to GenBank Accession numbers: EU443645-56 & GQ240942) (see Supplementary Fig. S1).
Ancient DNA
We successfully extracted and sequenced DNA from 34 out of 177 ancient domestic pig bones sampled from archaeological sites in Israel (Fig. 1). The overall success rate was 19%, which is the expected rate given the climate in the region38,39 (see Supplementary Fig. S2). The samples were collected from secure stratigraphic contexts associated with well-defined ceramic assemblages, which provide secure relative chronology (i.e., period and/or sub-period). Many of the Bronze and Iron Age strata from which the bones were obtained have been radiocarbon dated (e.g.40,41,42,43,44). We further obtained three dates out of the four samples we submitted for radiocarbon dating (there was insufficient collagen for dating in the fourth) (see Supplementary Table S1). Two fragments of the mtDNA CR were amplified: one (ANC1) following Larson et al.5, the second was chosen to increase resolution for the Near Eastern clade, and was targeted to the 3′ end of the modern CR fragment.
The ancient mtDNA data reveal the existence of two Near Eastern haplotypes in Israel in the past (Arm 1T and Y1) (Fig. 3). Haplotype Arm 1T is present in six of the 34 sequenced samples: one sample dating to the Middle Bronze I (MMP168 dated to 1895-1610 cal. BCE), three samples dating to the Iron I (11th century BCE), one to the Iron IIA (9th century BCE) and one to the Byzantine period (MMP253 dated to 400-605 cal. CE) (see Supplementary Table S1). Haplotype Y1 is present in the Late Bronze III to the Iron IIA (ca. 1200-800 BCE). The major turnover in the pig population occurred at the beginning of the Iron II, around 900 BCE, when the European haplotype first became predominant relative to the Near Eastern haplotypes (one sample is dated, MMP175 1005-805BC cal. BCE). One sample with a European haplotype comes from a Middle Bronze context (2000-1550 BCE) in the port city of Ashkelon. Unfortunately, there was insufficient soluble collagen to radiocarbon date this bone and confirm its age. The sample was re-extracted three times and multiple amplifications resulted in the same sequence.
In order to follow changes in the frequency of haplotypes through time and space, and understand the relationship between the samples, we created a three-dimensional statistical parsimony network of mtDNA CR (Fig. 4). The network is in agreement with the modern phylogenetic tree of Sus scrofa in which Europe, the Near East and East Asia fall into separate groups. The two Near Eastern haplotypes (Arm 1T and Y1) from this region are missing from modern Israeli pigs but are still found elsewhere in the Near East.
Layers represent the haplotypes in four different time periods. The top layer represents modern samples from all of Eurasia, while the three ancient layers represent ancient Israeli samples only. Each circle represents one haplotype. The colour corresponds to the different geographic regions: red-Europe; yellow-Y1 Near Eastern haplotype; pale blue - Arm 1T Near Eastern haplotype; pale green - other haplotypes from Near East; grey- East Asia. Small black dots represent missing haplotypes. The size of the circle is proportional to haplotype frequency. Vertical lines link haplotypes found in both time periods.
In the ancient Israeli specimens, we found additional mutations to the Near Eastern haplotypes Arm 1T and Y1: a mutation in the combined fragment in one Iron Age sample (MMP63), and an insertion in the Y1 haplotype in three samples from the Late Bronze and Iron I (MMP121, 129, 242). We also observed mutations in the European haplogroup in two specimens from the Roman period (MMP150, 211). These samples were re-extracted and multiple amplifications resulted in the same sequence. However, they are still confined to the general haplotypes that are of interest to us.
Discussion
Our results show that all modern Israeli wild boar populations share a European mitochondrial genetic signature (Fig. 2). This is despite the fact that the samples derive from different habitats–coastal, mountainous and desert, and represent a period of 60 years. Our results confirm those of Giuffra et al.3, who found a distinction between wild boars from Israel and those from the rest of the Near East, where local haplotypes are dominant in the modern populations5,6,7,9.
The ancient DNA data offer an explanation for this paradox. The data demonstrate that domestic pigs from the region possessed Near Eastern haplotypes (Arm 1T and Y1) (Fig. 3). These haplotypes were also found in Anatolian wild boars before domestication, starting in the Neolithic period, meaning that these haplotypes were the local genetic signature in the region (Fig. 5a). During the early phases of the Iron Age, however, domestic European pigs were introduced into the region. A millennium or so later, this genetic signature reached fixation in terms of mtDNA (Fig. 3). Subsequent introductions of European pigs into the southern Levant might have occurred after the Iron Age—for instance in Roman-Byzantine times and during the Crusades—and this may have contributed to the dominance of the European haplotype.
The number of samples in each period is indicated on the side of the haplotype frequency. Question marks refer to two undated samples. Wild boars are mainly represented in the Neolithic to Bronze period from Anatolia (a), and from the present day (g). Domestic pigs are mainly represented through the periods between (b–f). The maps were created using the ArcGIS version 9.3.1.
The introduced domestic European pigs had a crucial effect on the mtDNA of the indigenous wild boar populations of Israel. Feral European pigs could have driven the local wild boars to extinction, or, more likely, they could have hybridized the local wild boar population. Although the MC1R data show that modern wild boars from Israel possess a European wild boar coat colour (see Supplementary Fig. S1), we cannot distinguish whether this signal is a true local signature or whether it comes from the introduced European domestic pigs that went feral and retained the wild boar coat colour. Data regarding this gene from other parts of the Near East are missing and hence we cannot reach firmer conclusions.
The outcome of these introductions, in terms of mtDNA, was a complete turnover (Fig. 3). In Anatolia, a similar pattern is observed in the Late Bronze Age (samples dated to ca. 1,600-1,440 BCE), when European haplotypes first appeared. In Anatolia too the major transition to a European haplotype took place during the Iron Age9. But, in contrast to the situation in Israel, where the European haplotype became the dominant genetic signature, in Anatolia the modern wild boar populations still possess Near Eastern haplotypes9 (Fig. 5). In Israel this could have happened through repeated admixture with introduced European domestic pigs that went feral as well as severe population bottlenecks. A recent documented bottleneck that occurred during the 1930s and 1940s, when wild boar populations were decimated by overhunting19, may explain the lack of genetic diversity across the modern populations and the fixation of the European signature.
Although we cannot refute the possibility that European pigs were brought to the southern Levant in an earlier period (see the sample from Middle Bronze Ashkelon, but note that it could not be radiocarbon dated and may be intrusive), their first main appearance occurred during the Iron Age. The major shift between Near Eastern to European haplotypes in Israel took place around 900 BCE. This could have been connected to the migration and settlement of the Sea Peoples – groups that originated in the Aegean Basin, Cyprus and/or southern Anatolia. According to textual material and archaeological finds, in the 12th to early 10th centuries BCE, they settled along the coastal plain of the Levant. The Philistines are the best-known and most studied group among these peoples. They established themselves mainly in urban centres in the southern coastal plain, such as Gath (Tell es-Safi), Tel Miqne-Ekron and Ashkelon (e.g.30,31,33,45). The arrival of the Philistines was accompanied by a sharp rise in the ratio of pig bone in their main urban sites30,45. The exceptionally high frequencies (up to 20–25% of the faunal assemblage) stand in stark contrast to the low numbers of pigs in most contemporaneous assemblages from local Canaanite and early Israelite settlements (usually <2% of the faunal assemblage)23,29,30,46,47. Morphological evidence from Tel Miqne-Ekron also points to the possible entry of a new pig population into the site with the arrival of the Philistines48. Since the distribution of Philistine pottery indicates at least cultural diffusion to broader areas of the region, beyond the coast of the Levant, e.g., to the north and inland of the Levant, it is possible that the introduction of European pigs might have been a broader phenomenon. Note that later migrations from the west, for instance in the Hellenistic-to-Byzantine periods and the time of the Crusades, could have re-introduced European pigs into coastal areas as well as inland parts of the Levant.
The abundance of pig remains, together with the genetic results presented here, hint at the possibility that pigs were translocated by the Sea Peoples in general and the Philistines in particular during their migration to the southern Levant. A ca. 250 year gap exists between their first arrival and our first European mtDNA signal. This can be explained by several factors: 1) the Sea Peoples migration was a several-decades-long process (e.g.49) that seems to have peaked in the 11th century BCE; 2) most of our Iron Age samples come from non-Sea Peoples' sites; 3) the sample-size is too small to track minute chronological processes within the early phases of the Iron Age; 4) it took some time for the haplotype to become significant.
The results of this study show that modern DNA by itself cannot reflect the entire history of a species, and emphasize the effect of turnovers in misinterpreting phylogeographic patterns. In the past, people transported pigs over significant distances and this played an important role in shaping the phylogenetic patterns we observe today. Ancient DNA is a powerful tool that aids in deciphering species histories, since phylogeographic patterns are far more complex than surmised from present-day biogeography.
Methods
Sample collection
We examined a total of 38 bone, hair, skin and soft tissue samples of modern wild boars from Israeli museum collections (see Supplementary Table S1). The samples cover the entire biogeographic distribution of wild boars in the country, and all four populations as suggested in Davidowitz and Horwitz50 based on morphological characteristics.
We also obtained 177 samples of ancient pig bones from archaeological sites across Israel, spanning the Neolithic to Crusader periods (ca. 7000 BCE to 1300 CE) (Fig. 1, Supplementary Table S1). Most of the samples are of domestic pigs as evident from demographic and osteometric data. They derive from well-excavated sites with secure stratigraphic contexts, in many cases accompanied by multiple radiocarbon-dates (e.g.40,41,42,43,44). Four samples were directly radiocarbon dated at the Weizmann Institute, Rehovot, Israel. The dates throughout the text are stated as calibrated dates.
DNA extraction and amplification
Modern samples
DNA from bones was extracted according to a method modified from Yang et al.51. Specifically, about 50 mg of bone powder was incubated with 0.44 M EDTA (pH = 8) (AMRESCO, USA), 0.1 M urea, and 20 mg/ml proteinase K (AMRESCO, USA) overnight at 56°C. After decalcification and digestion, the supernatant was concentrated to about 100 μl using Vivaspin filter 3000 MWCO (Sartorius Stedim Biotech), and then directly purified using silica based spin columns (Minelute PCR Purification kit, QIAGEN, Inc).
DNA from tissue, skin, hair, and other non-bone samples were extracted using QIAamp DNA Micro Kit (QIAGEN, Inc) following a protocol for forensic casework samples.
Following Larson et al.5,7 a total of 620 bp of the Control Region (CR) of the mitochondrial DNA (mtDNA) was amplified using two overlapping fragments. The primers; PIG1F (5′-CATTCCATTCGTATGCAAACC-3′) and PIG3R (5′-ACCAGATGCCTGTTAAAGTT-3′) were used to amplify a 372 bp fragments5, and L119 5′-(CAGTCAACATGCATATCACC-3′) and H16108 (5′-GCACCTTGTTTGGATTRTCG-3′)7 amplified a 352 bp fragment.
In addition, following Li et al.37, a total of 1552 bp of the MC1R region was amplified. This region includes 425 bp of the 5′ untranslated region, 963 bp of the coding region and 164 bp of the 3′ untranslated region. One pair of primers was designed to amplify the whole region; MF1 (5′-GTGCGGCGGCTCTGCGCTCCAA-3′) and MR1 (5′-CCCCCACTCCCCATGCCTCCTG-3′)37. Another set of inner primers was designed for samples that were badly preserved and for which the amplification of the long fragment failed; MF2 (5′-GCTGCTGCTGGAGGCGGGC-3′), MR2 (5′-ACACCATGGAGCCGCAGATGAGC-3′).
Each PCR reaction contained a total volume of 25 μl, consisting of 1× PCR Buffer, 1-1.5 U of Platinum Taq DNA polymerase, 1.5–2 mM of MgCl2, 0.2 mM of each deoxynucleotide triphosphate (dNTP) (all from Invitrogen, UK), 0.1 mg/ml of Bovine Serum Albumin (New England, BioLabs, UK), 0.2–0.4 μM of each primer (Sigma-Aldrich Inc.), and, 2–3 μl of DNA extract. Reaction profiles included a 2-min initial denaturation step at 94°C, followed by 30–35 cycles of the following: 30 sec denaturation at 94°C, 30 sec annealing at 52–68°C, 0.5–1.5 min of extension at 72°C, and a final extension of 10 min at 72°C.
The amplicons were cleaned from unincorporated primers using Exonuclease I, and Shrimp Alkaline Phosphatase (Thermo Fisher Scientific, UK). The samples were sequenced and analysed in ABI 3100 Genetic Analyzer or Beckman-Coulter CEQT 8000 Genetic Analysis System. Sequencing was conducted on both strands. Sequencing chromatograms were assembled and analysed using analysis software (Geneious version 5.6 created by Biomatters. Available from http://www.geneious.com/).
Ancient samples
DNA extractions and preparations for PCR reactions of the ancient samples were set up at the Institute of Archaeology of Tel Aviv University (Israel) in a dedicated lab for ancient DNA, in a building where no molecular work has previously taken place or is currently being conducted.
The bone extraction procedure was similar to that used for modern samples. Following Larson et al.5, a short fragment of mtDNA CR was amplified ANC1F (5′- CTTTAAAACAAAAAAACCCATAAAAA-3′), and ANC1R (5′- TTAATGCACGACGTACATAGG-3′). This 74 bp fragment is highly variable and can distinguish between European, Near Eastern and East Asian haplotypes. In addition, in order to increase resolution between the European and Near Eastern haplotypes, another pair of primers was used at the 3′ end of modern fragment to amplify 79 bp of the mtDNA CR. The forward was designed for this study, namely ANC4 F (5′- GCACCTTGTTTGGATTRTCG), and the reverse primer was the same as in the modern samples H16108 (5′-GCACCTTGTTTGGATTRTCG-3′) 7.
PCRs and post-PCR work were performed at the Zoology Department of Tel Aviv University (Israel). PCR amplifications were performed in 25 μl reactions with: 1× PCR buffer, 1.5 U of Platinum Taq DNA polymerase High Fidelity, 2 mM of MgSO4, 0.2 mM of each dNTP (all Invitrogen, UK) 0.1 mg/ml of Bovine Serum Albumin (New England, BioLabs, UK), 0.4 μM of each primer (Sigma-Aldrich Inc.) and 2–4 μl of DNA extract. The PCR amplification consisted of an initial denaturation at 94°C for four minutes, followed by 55 cycles of denaturation at 94°C for 30 sec, annealing at 52°C for 30 sec, and extension at 68°C for 40 sec with a final extension period of 5 minutes at 68°C. The samples were cleaned, sequenced and analysed the same way as the modern samples.
Authenticity criteria for the ancient DNA data
For both modern and ancient samples multiple negative controls were used during DNA extractions (every fourth or fifth sample) and in all PCR reactions. All reagents used were molecular biology grade, and when possible, were decontaminated using UV irradiation. All working areas and equipment were decontaminated using bleach, and/or UV irradiation.
All extracts were independently amplified and sequenced at least twice for each fragment to ensure that there were no misincorporated bases. In addition, two thirds of all ancient samples were re-extracted and the two fragments were amplified. For the ANC1 fragment, another forward primer was designed, to insure repeatability. The primer ANC2F (5′- AAATTGCGCACAAACATACAAAT-3′) amplifies a shorter fragment (54 bp), and was used to re-amplify 80% of the samples.
For more details about the archaeological sites information, DNA success ratio and radiocarbon sample pre-treatment and measurement, please see Supporting Information.
Phylogenetic analyses
Modern samples
The modern DNA sequence data for mtDNA CR and MC1R were aligned with modern pig sequences available on GenBank (Supplementary Tables S1 and S2).
Phylogenetic relationships for mtDNA CR region were estimated using Maximum Likelihood (ML)52, and Bayesian algorithms53,54. jMODELTEST version 2.155 was used to determine which molecular substitution model best fitted the data based on the Bayesian Information Criterion. The chosen model was implemented in ML and Bayesian inference. ML phylogeny was estimated using PAUP* 4.0b1056. A full heuristic search was performed starting with a Neighbour Joining tree and tree-bisection-reconnection (TBR) branch swapping. The parameter values identified by jMODELTEST were used to construct the first tree. The parameters were then re-estimated from the resulting tree and used in a new ML analysis. The procedure was repeated until the estimated parameter values remained unchanged, and these values were used for 100 bootstrap replicates. MrBayes v3.2.153,54 was used to conduct the Bayesian Markov Chain Monte Carlo (MCMC) phylogenetic inference using best-fit model indicated by jMODELTEST version 2.155 based on the Akaike and Bayesian Information Criterion. The molecular substitution model was Hasegawa-Kishino-Yano (HKY)57 with gamma-distribution and a portion of Invariable sites to account for among site rate variation. MCMC sampling was performed using two separate runs of three million generations with sampling every 100 generations, discarding the first 10% as burn-in. Four chains (three heated, one cold) were used in the analysis in each of the two independent runs. Mixing and convergence to stationary distributions were checked with Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/). The desert Warthog (Phacochoerus aethiopicus) served as an outgroup.
Network software version 4.6.1.10 (www.fluxus-engineering.com) was used to construct median-joining network for MC1R region (Supplementary Table S1, Table S2)58. To be able to use the large dataset from Fang et al.36, the analysis for the creation of the network was carried out for 963 bp, which was mainly based on the coding region of MC1R.
Ancient samples
The two mtDNA CR fragments amplified for ancient DNA were concatenated and aligned with modern sequences from across Eurasia, given the large dataset that has been generated by previous studies5,6,9. Haplotype name for each specimen was based on the first fragment- ANC1 terminology.
Thirty-four ancient samples and 179 modern Sus scrofa sequences (including the 25 Israeli wild boars) were used to construct a 3D statistical parsimony network using the script TempNet59 in R60 (Supplementary Table S1, Table S2). Gaps were assigned as a fifth character. Using the Bayesian phylogenetic analysis, samples were assigned to one of four time categories: modern, Hellenistic to Crusader (330 BCE – 1200/1300 CE), Iron Age (1150-586 BCE) and Bronze Age (3500-1150 BCE). DNA sequences have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/), with accession numbers KF525706-KF525782.
References
Avise, J. C. et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annual Reviews of Ecology and Systematic 18, 489–522 (1987).
Avise, J. C. Phylogeography: The history and formation of species (Harvard University Press, Cambridge, MA, 2000).
Giuffra, E. et al. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154, 1785–91 (2000).
Haile, J., Larson, G., Owens, K., Dobney, K. & Shapiro, B. Ancient DNA typing of archaeological pig remains corroborates historical records. J. Archaeol. Sci. 37, 174–177 (2010).
Larson, G. et al. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc. Natl. Acad. Sci. USA 104, 15276–81 (2007).
Larson, G. et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc. Natl. Acad. Sci. USA 104, 4834–9 (2007).
Larson, G. et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307, 1618–21 (2005).
Alexandri, P. et al. The Balkans and the colonization of Europe: the post-glacial range expansion of the wild boar,. Sus scrofa. J. Biogeogr. 39, 713–723 (2012).
Ottoni, C. et al. Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics. Mol. Biol. Evol. 10.1093/molbev/mss261 (2013).
Hongo, H., Anezaki, T., Yamazaki, K., Takahashi, O. & Sugawara, H. Hunting or management? The status of Sus in the Jomon period in Japan. in Pigs And Humans 10,000 Years of Interaction (eds. Albarella, U., Dobney, K., Ervynck, A. & Rowley-Conwy, P.) 109–130 (Oxford University Press, Oxford, 2007).
Rowley-Conwy, P. & Dobney, K. Wild boar and domestic pigs in Mesolithic and Neolithic southern Scandinavia. in Pigs And Humans 10,000 Years Of Interaction (eds. Albarella, U., Dobney, K., Ervynck, A. & Rowley-Conwy, P.) 131–155 (Oxford University Pres, Oxford, 2007).
Vigne, J. D. et al. Pre-Neolithic wild boar management and introduction to Cyprus more than 11,400 years ago. Proc. Natl Acad. Sci. USA 106, 16135–38 (2009).
Hewitt, G. M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 68, 87–112 (1999).
Hewitt, G. M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 (1996).
Taberlet, P., Fumagalli, L., Wust-Saucy, A. G. & Cosson, J. F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 7, 453–464 (1998).
Hofreiter, M. et al. Lack of phylogeography in European mammals before the last glaciation. Proc. Natl. Acad. Sci. USA 101, 12963–12968 (2004).
Valdiosera, C. E. et al. Staying out in the cold: glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 16, 5140–5148 (2007).
Bray, S. C. E. et al. Ancient DNA identifies post-glacial recolonisation, not recent bottlenecks, as the primary driver of contemporary mtDNA phylogeography and diversity in Scandinavian brown bears. Diversity and Distributions 19, 245–256 (2012).
Mendelssohn, H. & Yom-Tov, Y. A report of birds and mammals which have increased their distribution and abundance in Israel due to human activity. Israel J. Zool. 45, 35–47 (1999).
Geraads, D. & Tchernov, E. Femurs humains du Pleistocene moyen de Gesher Benot Ya'acov (Israel). L'Anthropologie 87, 138–141 (1983).
Haber, A. & Dayan, T. Analyzing the process of domestication: Hagoshrim as a case study. J. Archaeol. Sci. 31, 1587–1601 (2004).
Marom, N. & Bar-Oz, G. The prey pathway: A regional history of cattle (Bos taurus) and pig (Sus scrofa) domestication in the northern Jordan Valley, Israel. PLoS One 8, e55958 (2013).
Hesse, B. Pig Lovers and Pig Haters: Patterns of Palestinian Pork Production. Journal of Ethnobiology 10, 195–225 (1990).
Grigson, C. Plough and Pasture in the early economy of the southern Levant. in The Archaeology of Society in the Holy Land (ed. Levy, T. E.) 245–68 (Leicester University Press, London, 1998).
Hesse, B. & Wapnish, P. Can pig remains be used for ethnic diagnosis in the Ancient Near East? in The Archaeology of Israel, Vol. 237, (eds. Silberman, A. S. & Small, D.) 238–270 (1996).
Hesse, B. & Wapnish, P. Pig use and abuse in the ancient Levant: Ethnoreligious boundary-building with swine. in Ancestors for the Pigs: Pigs in Prehistory. Vol. 15, (ed. Nelson, S. M.) 123–135 (1998).
Horwitz, L. K. & Studer, J. Pig production and exploitation during the Classical periods in the Southern Levant. in Archaeozoology of the Near East, Vol. VI, (eds. Buitenhuis, H., Choyke, A. M., Martin, L., Bartosiewicz, L. & Mashkour, M.) 222–239 (ARC-Publicaties 123, Groningen, 2005).
Lev-Tov, J. S. E. Pigs, Philistines, and the ancient animal economy of Ekron from Late Bronze to Iron Age II University of Tennessee. (2000).
Sapir-Hen, L., Bar-Oz, G., Gadot, Y. & Finkelstein, I. Pig husbandry in Iron Age Israel and Judah: New insights regarding the origin of the “Taboo”. Zeitschriftfirt des Deutschen Palästina-Vereins 129.1, 1–20 (2013).
Maeir, A. M., Hitchcock, L. A. & Horwitz, L. K. On the constitution and transformation of Philistine identity. Oxford J. Archaeology 32, 1–38 (2013).
Yasur-Landau, A. The Philistines and Aegean migration at the end of the Late Bronze Age (Cambridge University Press, Cambridge, 2010).
Kislev, M. E., Artzy, M. & Marcus, E. Import of an Aegean food plant to a Middle Bronze IIA coastal site in Israel. Levant 25, 145–154 (1993).
Oren, E. D. The Sea People and their world: A reassessment, (The University Museum, University of Pennsylvania Press Philadelphia, 2000).
Kijas, J. M. H. et al. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 150, 1177–1185 (1998).
Vage, D. I., Klungland, H., Lu, D. & Cone, R. D. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm. Genome 10, 39–43 (1999).
Fang, M. Y., Larson, G., Ribeiro, H. S., Li, N. & Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341 10.1371/journal.pgen.1000341 (2009).
Li, J. et al. Artificial selection of the melanocortin receptor 1 gene in Chinese domestic pigs during domestication. Heredity 105, 274–281 (2010).
Allentoft, M. E. et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. P. Roy. Soc. Lond. B Bio. 279, 4724–4733 (2012).
Bollongino, R., Tresseta, A. & Vignea, J. D. Environment and excavation: Pre-lab impacts on ancient DNA analyses. Comptes Rendus Palevol 7, 91–98 (2008).
Gilboa, A. & Sharon, I. An archaeological contribution to the Early Iron Age chronological debate: Alternative chronologies for Phoenicia and their effects on the Levant, Cyprus, and Greece. B. Am. Sch. Oriental Re. 332, 7–80 (2003).
Mazar, A., Bruins, H. J., Panitz-Cohen, N. & Van der Plicht, J. Ladder of time at Tel Rehov: Stratigraphy, archaeological context, pottery and radiocarbon dates. in The Bible and Radiocarbon Dating: Archaeology, Text and Science (eds. Levy, T. E. & Higham, T.) 193–255 (Equinox, London, 2005).
Boaretto, E. Radiocarbon Dates. in Megiddo IV: The 1998–2002 Seasons (eds. Finkelstein, I., Ussishkin, D. & Halpern, B.) 550–557 (Tel Aviv, 2006).
Sharon, I., Gilboa, A., Jull, T. A. J. & Boaretto, E. Report on the first stage of the Iron Age dating project in Israel: Supporting a low chronology. Radiocarbon 49, 1–46 (2007).
Finkelstein, I. & Piasetzky, E. Radiocarbon dating the Iron Age in the Levant: A bayesian model for six ceramic phases and six transitions. Antiquity 84, 374–385 (2010).
Hitchcock, L. A. & Maeir, A. M. Beyond Creolization and Hybridity: Entangled and Transcultural Identities in Philistia. Archaeological Review from Cambridge 28, 51–72 (2013).
Faust, A. & Lev-Tov, J. The constitution of Philistine identity: Ethnic dynamics in twelfth to tenth century Philistia. Oxford Journal of Archaeology 30, 13–31 (2011).
Bunimovitz, S. & Lederman, Z. Canaanite resistance: The Philistines and Beth-Shemesh - A case study from the Iron Age I. B. Am. Sch. Oriental Res. 364, 37–51 (2011).
Owen, J. The rise and fall of the Philistine pig: An investigation of a pig assemblage from Tel Miqne-Ekron. BSc Dissertation, University of Durham. (2005).
Cifola, B. The role of the Sea Peoples at the end of the Late Bronze Age: A reassessment of textual and archaeological evidence. Orientis Antiqvi Miscellanea I, 1–23 (1994).
Davidowitz, G. & Horwitz, L. K. Morphometric variation between populations of recent wild boar in Israel. in Pigs and Humans 10,000 Years of Interaction (eds. Albarella, U., Dobney, K., Ervynck, A. & Rowley-Conwy, P.) (Oxford University Press, Oxford, 2007).
Yang, D. Y., Eng, B., Waye, J. S., Dudar, J. C. & Saunders, S. R. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. of Phys. Anthropol. 105, 539–543 (1998).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–76 (1981).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Posada, D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–6 (2008).
Swofford, D. L. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4 (Sinauer Associates, Sunderland, Massachusetts, 2003).
Hasegawa, M., Kishino, H. & Yano, T. A. Dating of the human ape splitting by a molecular clock of mitochondrial-DNA. J. Mol. Evol. 22, 160–174 (1985).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Prost, S. & Anderson, C. N. K. TempNet: a method to display statistical parsimony networks for heterochronous DNA sequence data. Methods in Ecology and Evolution 2, 663–667 (2011).
Team, R. D. C. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2010).
Acknowledgements
We thank the following archaeologists, zooarchaeologists and museum curators for kindly providing us samples: Noha Agha, Amani Abu Hamid, Doron Ben-Ami, Daniel Berkowic, Fanny Bocquentin, Eric H. Cline, Ehud Galili, Ayelet Gilboa, Zvi Greenhut, Gila Kahila Bar Gal, Seymour Gitin, Amir Golani, Hamudi Khalaily, Thomas E. Levy, Nimrod Marom, Daniel Master, Assaf Nativ, Amihai Mazar, Ilan Sharon, Danny Syon, Assaf Yasur-Landau, Edna Stern, Karin Tamar, David Ussishkin, Shlomit Weksler-Bdolah. We also want to thank to the following museums for providing us samples: the Steinhardt National Collection of Natural History, Zoological Museum at Tel Aviv University (Israel), and the Wildlife Tissue Collection at the Laboratory of Molecular Evolution, National Natural History Collections of The Hebrew University of Jerusalem (Israel). This study was funded by the European Research Council under the European Community's Seventh Framework Program (FP7/2007–2013) ERC grant agreement no. 229418. Partial funding (relating to the excavation of the samples from Tell es-Safi/Gath and the research conducted by L.K.H) was provided by a grant (#32/11) from the F.I.R.S.T. (Bikura) track of the Israel Science Foundation to A.M.M. Ehud Weiss, and L.K.H. A small funding was also provided by the Jacob M. Alkow Chair in the Archaeology of Israel in the Bronze and Iron Ages.
Author information
Authors and Affiliations
Contributions
M.M., G.B.O., S.W. and I.F. designed research; M.M. performed research; M.M., D.H., G.L., S.W. and I.F. analysed data; G.B.O., L.K.H., A.M.M., L.S.H. and I.F. provided key samples and contextual information; E.B. performed the radiocarbon dating of the samples; M.M., D.H., G.B.O., G.L., S.W. and I.F. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (DOC 1589 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Meiri, M., Huchon, D., Bar-Oz, G. et al. Ancient DNA and Population Turnover in Southern Levantine Pigs- Signature of the Sea Peoples Migration?. Sci Rep 3, 3035 (2013). https://doi.org/10.1038/srep03035
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03035
This article is cited by
-
The Archaeology of Pig Domestication in Eurasia
Journal of Archaeological Research (2020)
-
Unlocking the origins and biology of domestic animals using ancient DNA and paleogenomics
BMC Biology (2019)
-
Long-term morphological changes and evolving human-pig relations in the northern Fertile Crescent from 11,000 to 2000 cal. bc
Archaeological and Anthropological Sciences (2019)
-
Like a pig out of water: seaborne spread of domestic pigs in Southern Italy and Sardinia during the Bronze and Iron Ages
Heredity (2017)
-
Eastern Mediterranean Mobility in the Bronze and Early Iron Ages: Inferences from Ancient DNA of Pigs and Cattle
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.