Abstract
Detection methods and data from molecular characterization of genetically modified (GM) events are needed by stakeholders of public risk assessors and regulators. Generally, the molecular characteristics of GM events are incomprehensively revealed by current approaches and biased towards detecting transformation vector derived sequences. GM events are classified based on available knowledge of the sequences of vectors and inserts (insert knowledge). Herein we present three insert knowledge-adapted approaches for characterization GM events (TT51-1 and T1c-19 rice as examples) based on paired-end re-sequencing with the advantages of comprehensiveness, accuracy and automation. The comprehensive molecular characteristics of two rice events were revealed with additional unintended insertions comparing with the results from PCR and Southern blotting. Comprehensive transgene characterization of TT51-1 and T1c-19 is shown to be independent of a priori knowledge of the insert and vector sequences employing the developed approaches. This provides an opportunity to identify and characterize also unknown GM events.
Similar content being viewed by others
Introduction
It is internationally agreed that genetically modified (GM) crops could be commercialized after thorough safety assessment and only if they are deemed safe1. Molecular characterization of transgene inserts at the chromosome level including the insert sequence, its localization, the number of inserts and its flanking sequences is essential for the safety assessment and labeling of GMOs2. Furthermore, transgene insertion is frequently associated with intended and unintended changes at the genomic, transcriptomic, proteomic and metabolomics levels, which potentially affects food/feed quality and safety3. Therefore, molecular characterization data on the complete insert sequences and their localization are particularly important both for developers, risk assessors and regulators of GM crops. These data also serve as a basis for the development and validation of the specific detection methods for GMOs monitoring4.
Current legally required and commonly applied approaches to obtain molecular characterization data are limited to Southern blot and polymerase chain reaction (PCR) analyses, combined with standard sequencing of the functional (intended) insert(s) and flanking genomic DNA of the host2. These approaches are time consuming and their abilities to detect DNA sequence motifs are potentially limited by various factors, such as the existence of substitutions, insertions, deletions in the sequence and the quantities and/or sizes of the targets. Thus their output information are often only sub-optimal albeit the optimal input efforts. For example, the developer initially documented only one inserted copy of the expression cassette of the 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) gene in the soybean event GTS40-3-2 (Roundup Ready, OECD unique identifier [UI] = MON-Ø4Ø32-6) that was approved for commercialization in 1994 5. Later, the rearrangement of the 3' NOS terminator junction and one unintended 70-base pair (bp) DNA fragment insertion were evidenced in this event and the molecular characterization of GTS40-3-2 has been amended three times6,7,8. With the constant expansion of GMO research and development, there is an increasing number of reports about fields or foods/feeds containing illegally/unknown released GMOs. Examples include but are not limited to the StarLink maize (CBH351, UI = ACS-ZMØØ4-3), the GM rice events LL601 (UI = BCS-OSØØ3-7), Kemingdao and Kefeng 6 and the FP967 flax (UI = CDC-FLØØ1-2) cases, which caused public concerns and international trade disruptions. In addition, authorities are confronted with the very difficult task to detect unknown GMOs for which no information is available10. With the development of high throughput next generation sequencing (NGS) technology, complete genome sequences can be obtained at high sequencing depth at reasonable costs11. NGS approaches have proven to be powerful tools for discovering gene fusion, re-arrangements, DNA insertion and structural variations in different animal and plant samples12,13,14,15,16, although the massive data processing is the challenge. De novo assembly of a large eukaryote genome is presently beyond the scope of transgene characterization, but this challenge may be mitigated in the foreseeable future. Importantly, the majority of crop plants for which transgenes are developed have already been extensively studied. More or less complete genome assemblies for these are or will soon be publicly available (NCBI Genome Resources, http://www.ncbi.nlm.nih.gov/genome; Beijing Genomics Institute, Plant research, http://www.ldl.genomics.cn/page/pa-plant.jsp). Compared to conventional transgene characterization, whole genome re-sequencing, targeted bioinformatics analyses and limited de novo assembly emerges as a much simpler and more effective approach to transgene characterization.
Herein we present approaches to further exploit DNA re-sequencing and bioinformatics to comprehensively characterize the inserts of GMOs also when the a priori (pre-existing) knowledge of the DNA sequence(s) of vectors and inserts is limited or even absent. Rice is one of the most important crops in the world and a staple food for a large share of humanity. The complete sequence of the relatively small (389 Mb) rice genome was the first crop genome to be published17. Since our research interests are transgene characterization and detection and rice genetics, we chose the two aforementioned transgenic rice events, TT51-1 and T1c-19, as examples in this study. The TT51-1 event is the first food crop that was approved for commercialization in China in 200918,19 and the T1c-19 event is in the pipeline for approval in China20. We also included an in silico mimic to validate the software for detection of unknown transgenes.
Results
Three bioinformatics modules and the analytical program adapted to GMOs of different classes of pre-existing insert knowledge
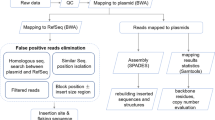
Since the knowledge of a GM available a priori is case dependent, we designed three different bioinformatics modules for data analysis ( Fig. 1 ). Each is targeted to be fit for a given hypothetical scenario (cf. insert sequence knowledge [ISK] classes 2–4 in the report of Holst-Jensen et al.)9. Module 1 is intended for use when the DNA sequence of the transformation vector is known (ISK-class 2). Module 2 is intended for use when a DNA sequence database of genetic elements and transgene constructs from known GMOs is available and can be used as a reference library (ISK-class 3). Module 3 is intended for use when no knowledge of the DNA sequence of the vector and insert is available a priori (ISK-class 4 = unknown GMO). For application of any of the three modules the species reference genome sequence must be available as a reference.
The three modules proposed for molecular characterization of transgenic lines using paired-end whole genome re-sequencing and data analysis.
Module 1 (Fig. 1a–1b): the complete DNA sequence of the transformation vector is available (corresponding to insert sequence knowledge [ISK] class 2 scenarios). The paired-end reads are characterized into five types (A to E). Type A: both paired-ends perfectly map back to the host genome. Type B: one end matches to the host genome, the other to the transgene. Type C: both paired-ends match to transgene. Types D and E: one end matches to host genome or transgene and the other spans the junction region between host genome and transgene. Module 2 (Fig. 1c): the DNA sequence of the transformation vector is not available but the transgene insert is expected to contain at least one genetic element that is included in a transgene element sequence library (database; corresponding to ISK-class 3 scenarios). Successful detection and characterization of the transgene depends on matches between the transgene and the sequence library. Module 3 (Fig. 1d): no DNA sequence information on the transgene insert is available and a transgene element sequence library is expected to be of limited use (cf. ISK-class 4). Successful detection and characterization depends on efficient de novo assembly and contig analyses.
Initially, the whole genomic DNA is isolated and subjected to standardized paired-end sequencing. The paired-end reads are then grouped according to their mapped affinity to known reference sequences (A to E; Fig. 1a ).
In module 1 ( Fig. 1b ), four consecutive steps are included as follows: i) After processing the raw data (including filtering and adaptor trimming), all NGS reads are mapped back to the host genome sequence to identify paired-end reads of type A or putatively to the types B, D and E; ii) Then reads not classified as type A are mapped to the known transgenic vector sequence to assign reads to types B, C, D or E); iii) The transgene integration site(s), number of inserts and flanking sequences are then determined by analysis of the type B, C, D and E reads; iv) Finally, the insert is verified using common PCR and Sanger sequencing analysis.
In module 2 ( Fig. 1c ), five steps are involved: i) Construction of a DNA transgene sequence library including frequently used exogenous genes, regulatory elements, marker genes and vectors from different open sources (publications, patents and available databases); ii) Mapping all reads back to the transgene sequence library; iii) Individual de novo assembly of matched reads and their paired reads; iv) Uncovering the transgene sequences of the sequenced sample and drawing of the sketch map of DNA insert(s) on the basis of the assembled contigs; v) Experimental confirmation of the transgenic inserts by conventional PCR and Sanger sequencing. Notably, there is no separate mapping to identify reads of type A prior to the de novo assembly in step iii. This is to ensure comprehensive transgenic insert retrieval also in cases where cis-genic (host-derived) elements are inserted. If type A reads are removed from the set of reads to be assembled, this could improve the data processing time at the cost of cis-gene elements detection.
In module 3 ( Fig. 1d ), four steps are involved: i) Mapping of the reads against the reference genome to subtract type A reads; ii) direct de novo assembly of remaining reads; iii) BLAST analysis of all inferred contigs; iv) experimental verification of inserts by conventional PCR and Sanger sequencing.
In order to simplify the workload of these developed modules in massive data analysis, we then developed one integrated (semi-) automated program to implement the three modules based on the basic principles of Burrows-Wheeler Aligner (BWA) and Assembly by Short Sequences (ABySS) algorithms21,22. It circumvents the need for multiple runs of step-by-step DNA alignment and de novo analysis to identify matching read-pairs and assemble putative insert contigs. The developed program is downloadable at the following URL: http://gmdd.shgmo.org/Computational-Biology/Transeq/Transeq.tar.bz2.
The characterization of GM rice events T1c-19 and TT51-1 employing the three modules
In order to confirm the applicability of the developed modules for characterization of biotech events two presumed representative transgenic rice events, T1c-19 and TT51-1, were selected as case examples for further analysis. The event T1c-19 was transformed with the cloning vector pBar-1C ( Supplemental sequence No. 1 ) into the rice cultivar Minghui 63 by Agrobacterium-mediated transformation. The event TT51-1 was co-transformed with two cloning vectors pFHBT1 ( Supplemental sequence No. 2 ) and pGL2RC7 ( Supplemental sequence No. 3 ) into Minghui 63 by particle bombardment. Paired-end sequencing (90 bp reads) yielded 8.97 Gb and 9.92 Gb of raw sequence data corresponding to approximately 23.8 × and 26.4 × sequencing depth from T1c-19 and TT51-1 rice, respectively. The raw sequence reads of T1c-19 and TT51-1 rice are available in the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) with accession number SRA057974. The raw data was imported into the developed bioinformatics program for further analysis with the three different modules.
T1c-19 rice
Using the module 1 ( Fig. 1b ), ~ 75.4 million (75.76%) reads were attributed to the type A, both ends properly mapped to the rice reference genome (TIGR 7.0), while 2,613 reads were type C, matching to the transgenic vector pBar-1C ( Table 1 ) and 128 chimeric pairs of reads were Type B, D and E, under the filter parameters allowing maximum edit distance (including insertions, deletions or substitutions) 10 bp for each single read ( Supplemental Table S1 ). A total of 111 pairs of reads were perfectly matched to rice chromosome (Chr) 11 and pBar-1C and 10 pairs of reads were perfectly mapped to rice Chr 04 and pBar-1C. These 121 pairs of reads are compatible with presence of two transgene inserts located on Chr 04 and Chr 11, respectively. The flanking sequences of these two transgene insertions were also obtained. Two pairs of reads mapped to Chr 12 and were regarded as false positive due to the repetitive genome sequence between the Chr 12 and Chr 11. The left five pairs of scattered reads mapped to other chromosomes (Chr 01, Chr 10 and Chr 05) were observed and regarded as artefacts because of sequence and mapping specificity ( Supplemental Table S1 ). Relaxing the stringency of the mapping parameters only slightly affected the number of chimeric read-pairs ( Supplemental Table S1 ). In order to validate the above mentioned results, thirteen primer pairs were designed based on the obtained 128 paired reads ( Supplemental Table S2 ). PCR was performed with these primers employing the T1c-19 rice and its isogenic control line Minghui 63 as templates, which led us to conclude that except for the Chr 04 and Chr 11 matches, all matches were artefacts and did not represent additional inserts or transgene rearrangements ( Fig. 2a, b ). Supportively, the long distance PCR amplification and Sanger sequencing results confirmed the above two transgene insertions. A 6394 bp and a 6392 bp T-DNA insertions were found to be inserted at 31,763,777 on Chr 04 and at 1,124,835 on Chr 11, respectively ( Supplemental sequence No. 4 and No. 5 ). The sequence alignment analysis authenticated the two intact T-DNA insertions in the rice host genome (cf. Fig. 3a ). To check for putative integration of transgene vector backbone sequence, a coverage depth plot of reads mapped against the pBar-1C vector was prepared ( Supplemental Fig. S1 ). No indications of vector backbone sequence insertion were observed in T1c-19 rice. The number of T-DNA inserts was also verified using equation 1 (see details in Materials and Methods section). The inferred insert detection index (X) was 1.02 (≈1.0), indicating that all inserts derived from the transformation vector were retrieved ( Table 1 ). Altogether, these results suggest the feasibility of the developed strategy (module 1 plus program) in the comprehensive molecular characterization of the GMO using re-sequencing. Under ISK-class 2 conditions9, we conclude that the input effort vs. output information ratio of the approach presented here is superior to the currently applied and legally required approaches including Southern blot and PCR analyses.
Comparison of inferred transgene maps for the rice events T1c-19 (Figs. 3a–3c and TT51-1 (Figs. 3d–3f) and for the mimic insert spiked in silico into the T1c-19 rice event raw sequence data reads (Figs. 3g–3h).
Top: the correct maps, obtained with module 1 for the two rice events (Figs. 3a and 3d) and spiked in silico (Fig. 3g). Middle: the results inferred with module 2 for the two rice events (Figs. 3b and 3e). Bottom: the results inferred with module 3 for the two rice events (Figs. 3c and 3f) and the spike (Fig. 3h).
In the analysis with module 2 ( Fig. 1c ), the rice T1c-19 was treated as a blind (unknown) sample. A transgene element library was constructed containing the DNA sequence of 134 elements frequently used in GMOs (promoters, terminators, genes, cloning vector elements) retrieved from published literature, patents and databases ( Supplemental Table S3 ). A total of 29,861 pairs of reads ( Supplemental Table S4 ) mapped to 25 elements in the constructed library, indicating the presence of transgene(s), such as the maize Ubiquitin promoter, CaMV35S promoter, CaMV35S polyA terminator, NOS terminator and the Bar gene. De novo assembly of the 29,861 pairs reads yielded 22 contigs with length > 90 bp ( Supplemental Table S5 ). BLASTN analysis of these contigs against the GenBank database showed the presence of several genetic elements frequently used in transgene constructions (maize Ubiquitin promoter, CaMV35S promoter, CaMV35S PolyA terminator, NOS terminator, Lac Z, Cry1C gene and Bar gene), rice endogenous genes (Sucrose Phosphate Synthase gene, Acetyl-CoA carboxylase gene and Actin promoter) and the insert junctions on Chr 11 ( Supplemental Table S5 ). Notably, the Cry1C gene was not included in the transgene element library but was also among the observed putative insert elements of assembled contigs. The insert-junctions on Chr 04 detected in module 1 analysis were not identified in this approach. A sketch map of the transgene insert(s) was inferred (as shown in Fig. 3b ) and successfully confirmed with PCR and Sanger sequencing ( Supplemental sequences No. 4 and 5 ). The results confirmed the feasibility and practicability of module 2 for characterization and identification of GMOs referable to ISK-class 3 9, although its success depends, to some extent, on the constructed GMO library. The more complete the transgene element library, the more powerful the application of module 2. We are continuously improving the transgene element library and an update relative to the present version is available upon request.
The module 3 pipeline ( Fig. 1d ) was developed to deal with unknown GMOs that correspond to ISK-class 4 scenarios9, in which the information on the transgenic insertion is not available and the constructed GMO library cannot be used either. A total of 386,520 contigs (> 100 bp in length) were de novo assembled from all non-type A reads using the developed semi-automated program. The number and the length of assembled contigs are plotted in Supplemental Fig. S2 . BLASTN analysis of all contigs demonstrated that one 6,190 bp contig ( Fig. 3c ; Supplemental Sequence No. 6 ) had high similarity to the frequently used transgenic elements in transformation vectors (CaMV35S promoter and terminator, maize Ubiquitin promoter and NOS terminator) and exogenous genes (Cry1C, Bar and LacZ genes; as shown in Supplemental Fig. S3 ). We then compared the 6,190 bp contig with the known transgene T-DNA insert (cf. module 1 analysis above) and found that they were highly similar. This indicates that the module 3 pipeline is a very effective alternative for the molecular characterization of GMOs from ISK-class 4 scenarios except for the slightly large workload for blasting and filtering the assembled contigs (here 386,520).
TT51-1 rice
Using the module 1 ( Fig. 1b ), around 85.3 million (77.32%) reads were type A and 2,428 reads were type C, matching to the cloning vectors pFHBT1 and pGL2RC7 ( Table 1 ). In addition, 228 chimeric reads were Type B, D, or E, including 81 pairs on Chr 03, 45 pairs on Chr 10, 38 pairs on Chr 05, 18 pairs on Chr sy, 13 pairs on Chr 04, 7 pairs on Chr 09, 6 pairs on Chr 08, 5 pairs on Chr 06, 5 pairs on Chr 11, 3 pairs on Chr 02, 3 pairs on Chr 12, 2 pairs on Chr 07, 1 pair on Chr 1 and 1 pair on Chr un ( Supplemental Table S6 ). All these obtained paired reads were grouped based on the associated chromosome number and the paired reads of each group were used to reveal the transgene insertion sites and flanking sequences individually. The results showed that two transgene insertions occurred in TT51-1 rice, one located at position 2,640,325 on Chr 04 and another located at position 5,697,885 on Chr 10. The 81 pairs of reads on Chr 03 were obtained because the rice endogenous Actin promoter is present in both the rice genome and the T-DNA inserted of the vector pFHBT1. Other scattered reads that were mapped to other chromosomes (Chr 01, Chr 02, Chr 06, Chr 07, Chr 08, Chr 09, Chr 11, Chr 12, Chr sy and Chy un) were also observed and regarded as artefacts, after considering sequence and mapping specificity or the homogenous or repeat sequences among the 12 chromosomes in the rice genome ( Supplemental Table S5 ). Relaxing the stringency of the mapping parameters only slightly affected the number of chimeric read-pairs ( Supplemental Table S5 ). In order to confirm the above results, twenty primer pairs were designed based on the obtained 228 pairs of reads ( Supplemental Table S2 ). PCR was performed with these primers employing the TT51-1 rice and its isogenic control line Minghui 63 as templates. The PCR and sequencing results indicated that except for the Chr 04 and Chr 10 matches, all matches were artefacts and did not represent additional insertions or transgene rearrangements ( Fig. 4a, b ). PCR amplification and Sanger sequencing results confirmed the above two transgene inserts and provided details on the integrated T-DNA sequences. On Chr 10, one long (9,818 bp) tandem Cry1Ab/c cassette derived mainly from pFHBT1 was found to be integrated into the rice genome at position 5,697,885. It included partial rActin promoter, two tandem Cry1Ab/c cassettes and partial hpt gene ( Fig. 3d ; Supplemental sequence No. 7 ). This large insert was compatible with rearranged integration of one minor pGL2RC7 derived motif and two major and three minor partial pFHBT1 derived contigs into one large insert. On Chr 04, one short transgene insert (599 bp) consisting of partial rActin promoter and Cry1Ab/c gene derived from pFHBT1 was found to be integrated at position 2,640,325 ( Fig. 3d ; Supplemental sequence No. 8 ). To get an overview of which parts of the cloning vectors that apparently had been integrated into TT51-1 rice, coverage plots of reads mapped against the pFHBT1 and pGL2RC7 vectors were prepared Supplemental Fig. S4 and S5 ). These plots indicated the presence of almost all vector elements, except the CaMV35S promoter and hpt gene from the pGL2RC7 vector. Residual backbone sequences (bla and hpt gene and multiple cloning sites) were located to the Chr 10 insertion sequence ( Supplemental sequence No. 7 ). The number of T-DNA inserts was verified using equation 1 (see Materials and methods) and the inferred insert detection index (X) was 1.027 (≈1.0), indicating that all inserts derived from the transformation vectors were retrieved ( Table 1 ). Altogether these results suggest that we have provided a full molecular characterization of TT51-1 rice, with significant improvements compared to previous reports on TT51-1 rice18,23. As described above for T1c-19 rice, re-sequencing combined with the bioinformatics analysis with module 1 provides a novel approach for effective and comprehensive molecular characterization of GMOs.
In the analysis with module 2 ( Fig. 1c ), the rice TT51-1 was treated as a blind (unknown) sample. A total of 11,157 pairs of reads ( Supplemental Table S7 ) mapped to 14 elements in the constructed transgene element library, indicating the putative presence of transgene(s), such as the NOS terminator, bla gene, Cry1Ab/c gene and hpt gene. De novo assembly of the altogether 11,157 pairs reads yielded 12 contigs with the length > 90 bp. BLASTN analysis of these contigs against the GenBank database showed the presence of several frequently used transgenes and elements (NOS terminator, Cry1Ab/c gene, bla gene and hpt gene), rice endogenous genes (Sucrose Phosphate Synthase gene and Acetyl-CoA carboxylase gene) and two insert junctions on Chr 10 that were also retrieved with module 1 ( Supplemental Table S8 ). The insert junctions on Chr 04 (cf. module 1 analysis above) were not identified with this approach. A sketch map of the transgene insertions was inferred (as shown in Fig. 3e ) and successively confirmed with PCR and Sanger sequencing. As with T1c-19 rice, the results confirmed the feasibility and practicability of re-sequencing combined with bioinformatics analysis with module 2 for characterization and identification of GMOs referable to ISK-class 3.
For the module 3 pipeline ( Fig. 1d ) the scenario is of ISK-class 4 type (unknown GMO). From the set of non-type A reads, a total of 212,515 contigs (> 100 bp in length) were obtained by de novo assembly. The number and the length of assembled contigs are plotted in Supplemental Fig. S6 . BLASTN analysis of all contigs demonstrated that two contigs have high similarity to frequently used transgenic elements of transformation vectors. As was the case for T1c-19 rice also TT51-1 rice therefore does not meet the module 3 assumption that the transgene does not contain elements of the transgene element library for module 2. We will come back to this in the discussion below. One contig of 3,454 bp in length ( Supplemental Sequence No. 9 ), included the transgene elements of Cry1Ab/c gene, NOS terminator and bla gene flanked by or including the rice Actin promoter ( Supplemental Fig. S7 ). The other contig of 1,478 bp ( Supplemental Sequence No. 10 ) included a putative insert junction on Chr 10, corresponding to the 3′ end of the insert detected with modules 1 and 2 (see above). Furthermore, the 1,478 bp contig included the commonly used cloning vector backbone sequences of the bla and hpt genes ( Supplemental Fig. S8 ). Sketch maps of the transgene insertions were also drawn for the 3,454 and 1,478 bp contigs (as shown in Fig. 3f ). According to the sketch maps and verified sequences of TT51-1 insert with the two used cloning vectors (pFHBT1 and pGL2RC7), we confirmed that the rearrangement and truncation took place in the transformation process of particle bombardment24. As with T1c-19 rice, the results also indicate that the module 3 pipeline is a very effective alternative for GMOs in the ISK-class 4 scenarios.
The mimic transgene insertions from an artificial data set
To further confirm the ability of the developed module 3 to detect and characterize unknown transgene insertions, a third, artificial (simulated) dataset was created by in silico spiking of the T1c-19 dataset with mimicked reads corresponding to a 6424 bp contig containing one long (5,191 bp) and one short (89 bp) insert of “unknown” GM origin in the rice SPS gene ( Fig. 3g ; Supplemental Table S9; Supplemental Sequence No. 11 ). The “unknown” GM inserts of this mimic were almost entirely derived from soybean and the mimic did not contain any of the elements included in the constructed transgene element library or any cloning vector backbone elements.
For the analysis of this data set no information on the hypothetical cloning vector used was available. Thus it would not be possible to use module 1 for the analysis. In a real life situation, it is not possible to exclude a priori the possibility that the data set includes one or more of the elements in the transgene library. Thus it is reasonable to use module 2 for the analysis. However, in this simulated case it was known a priori that module 2 would not yield true positives and module 2 was not explored.
With module 3, the spiked data set was input into the semi-automated program and a total of 387,518 assembled contigs (> 100 bp in size) were generated. Comparing with the 386,520 contigs from T1c-19 rice data set (module 3 analysis above), more than 1,000 novel contigs were created. The BLASTN analysis of these new contigs revealed that two contigs of 5,200 bp and 238 bp, respectively ( Supplemental Sequence No. 12–13 ), had high similarities to insertions into the rice SPS gene of DNA from soybean and other plant species. Successively, we compared the sketch maps of the two contigs with the mimicked insert. As illustrated in Fig. 3h , these contigs closely resembled the mimicked insertion. Detailed alignment of the 5,200 bp contig with the simulated unknown insertion revealed that 4,939 bp of the contig mapped to the 6,424 bp mimic with 99.96% identity, including the two junctions between the rice SPS sequence and the insert. However, a 159 bp deletion within this insert was incorrectly indicated in the 5,200 bp contig. Similar alignment of the 238 bp contig revealed that 89 bp of the contig completely mapped to the 89 bp mimic with 100% identity and the two junctions between the rice SPS sequence and 89 bp mimic were also included. Combined with the two contigs of 5,200 bp and 238 bp, four short deletions were observed, i.e. the deletions of positions 1–418, 5358–5515, 5727–5902 and 6141–6424 of the mimic. However, these four deletions did not significantly affect the ability to characterize the transgene insertions. Concluding from the above results, the two mimic inserts were successfully retrieved with the pipeline of module 3 and the semi-automatic program. Notably one insert was as short as 89 bp. This should serve to demonstrate the power of the developed module 3 for the characterization of ISK-class 4 transgene inserts (unknown GMOs).
Discussion
The purpose of this study was to demonstrate the potential of high throughput sequencing for molecular characterization of GMOs under highly diverse a priori knowledge conditions. For this purpose three different bioinformatics modules were designed, each fit for a given insert-sequence knowledge class9. The results obtained from the combined analysis with all three modules on T1c-19 rice indicate that this rice event contains only two, nearly identical, large complete inserts. The molecular characterization of these two inserts and their insertion loci in T1c-19 rice are complete (module 1), nearly complete (module 2) or partial but sufficient to facilitate complete characterization (module 3). Notably, only one transgene insert ( Supplemental sequence No. 4 ) was revealed for the event T1c-19 using state-of-the-art Southern blot and PCR techniques according to a published report20 and cloned sequence (Genbank accession HQ161062.1). The results obtained with the combined use of all three modules on TT51-1 rice indicate that this rice event contains two inserts. The molecular characterization of the two inserts and their insertion loci in TT51-1 rice are complete (module 1), nearly complete (module 2) or partial but sufficient to facilitate complete characterization (module 3). One of the inserts in TT51-1 is short (599 bp) and unintended and has not been previously reported. The other is large (9818 bp) and rearranged. The partial sequence of the 9818 bp insert has previously been reported23. However, significant errors in the previous description of the 5′-end of the large insert were observed in the present study. The corrected full-length insert map perfectly integrates two previously published and apparently unlinked accessions HQ161054 (5′ flank/insert) and HQ161055 (3′ flank/insert). Obviously, the approaches reported here could be used to characterize GMOs at any ISK-levels.
Module 1 proved to be extremely effective for the comprehensive detection and characterization of the transgene insertions (sequences of inserts, insertion loci and flanking sequences and the number of inserts). It revealed one small insert (partial, truncated construct) and the complex rearranged long insert of TT51-1 rice. Module 1 may be particularly useful for transgene developers wishing to obtain complete molecular data for further selection among related transgene events, for safety assessments and for development of reliable event-specific detection methods.
Module 2 provides a basis for rapid complete characterization of event specific inserts and flanking sequences. It has the potential to serve as an alternative GMO-screening approach, similar to but more advanced than the commonly applied so-called matrix approach9. Module 2 may be particularly useful to agricultural supply chain stakeholders in need of knowing if their products contain authorized or un-authorized GMOs.
Module 3 lays a foundation for complete characterization of event specific insertions and flanking sequences of unknown GMOs and its suitability and validity has been demonstrated by the analyses of two GM rice events and mimic transgene insertion data. It is potentially of great value to public risk assessors and regulators for unknown GMOs inspection and monitoring.
With the two investigated rice events T1c-19 and TT51-1 the inserts contained several of the elements of our transgene element library. The contigs identified as the true inserts with the BLAST analyses were easily identified as transgenic due to their similarity to such elements. It could therefore be argued that these two rice events are unfit to assess the performance of module 3. We acknowledge this weakness associated with these two specific examples. However, the situation with the mimic transgene insertion was completely different. The transgenic insert of the mimic was entirely composed of sequence elements which are absent in the transgene element library. This example therefore demonstrates that module 3 is valid.
For (semi-) automated analysis with module 3, several options will be explored and exploited to increase the success rate and efficiency of transgene insert detection/identification in future versions of the software. The contig size distributions suggest that the contigs of interest are usually among the longest contigs. With module 3 the a priori information on the transgene is absent or extremely limited. Complete insert detection and characterization may therefore be less important at this stage than is the case with GMOs that can be characterized with modules 1 or 2. Examining the longest contigs first and perhaps more carefully than the shorter contigs can therefore be a reasonable approach, although it may result in failure to detect shorter transgenic inserts which cannot be represented by long contigs. Depending on the outcome of the BLAST analysis, there are alternative routes to transgene contig identification. Transgenes are by definition insertions of sequences from other taxa into the recipient genome. Thus, transgene inserts are likely to appear as contigs showing substantial dissimilarity to the reference genome (including chloroplast and mitochondrial genomes). The (main) transgene insert of a GMO is usually transcriptionally functional and includes a coding element. The coding element in many GMOs is derived from cDNA and therefore lacks introns. Such a transgene would contain a relatively long open reading frame (ORF) compared to endogenous ORFs. Screening all the contigs for long ORFs, starting with the longest contigs could therefore contribute to faster identification of the transgene contig(s). This approach would work independently of the result of BLAST analysis. It is, however, unfit for detection of genes containing introns. If the BLAST analysis returns annotated hits then another option emerges. The complete functional insert will normally include a promoter and usually also a terminator. This allows to study if a contig corresponds to a combination of promoter, gene and/or terminator elements. Notably and essentially by default, none of the elements found in the contig with the module 3 are included in the transgene element library of module 2. Verification of the identity of transgene contigs after the bioinformatics analysis should in all cases be done experimentally, e.g. using conventional PCR and DNA sequencing.
The re-sequencing and bioinformatics analyses outlined here are intended for samples derived from a single GM cultivar or species. The complexity of processing the combination of raw sequence data and necessary reference database accessions can be computationally prohibitive. This would particularly be true if a product containing ingredients of different species is analyzed. Intragenic inserts, if used, will be exceptionally difficult to detect without a priori knowledge of the DNA sequence of the transformation vector. We anticipate that increasing the sequence coverage can mitigate this challenge for modules 1 and partially also for module 2, but not for module 3. If a sufficiently large number of annotated reference cultivars are available this can also to some degree improve the prospects for module 3.
With module 1 the potential for detection and characterization of rearrangements of the recipient genome resulting from transformation can be maximized by re-sequencing the non-GM isogenic cultivar in paralleled. This is, however, not a prerequisite for successful comprehensive detection and characterization of inserts and flanking sequences, as demonstrated in this study.
Sequencing depth is positively correlated with the quality and quantity of data. The approaches outlined here can be further improved. Expansion of the transgene element library can improve the output of analyses with module 2, although for the two rice events examined in detail here the library was sufficiently complete to allow for complete insert characterizations. As discussed above, there is considerable potential for improvements of module 3 as well. The bioinformatics workload still represents a limitation to the routine use of modules 2 and 3 until the automatic and simplified software be developed in future.
The two raw-data sets obtained from T1c-19 and TT51-1 rice, respectively, are barely larger than the size of single data sets that can be obtained with the Illumina MiSeq sequencer. This fact is important because it suggests that it is possible to obtain the raw sequence data from a DNA sample within 1–2 working days with the new sequencer (Illumina MiSeq), as compared to 14 working days with the Illumina HiSeq sequencer used in the present study. Under optimal conditions we estimate that the total time required from receipt of sample material to presenting a detailed draft map of the insert (module 1) can be as little as three working days for a GMO with a small genome such as rice. Similarly, we estimate that without any a priori knowledge of the transformation vector or insert (module 3) a draft map of the insert excluding experimental verification can be obtained in as little as five working days with the help of automatic software in future. This would undoubtedly represent a significant improvement when time is critical, e.g. to manage risks and minimize damage from presence of unknown GMO in the supply chain.
Methods
GM rice samples
The transgenic rice T1c-19 was transformed with plasmid pBar-1C ( Supplemental sequence No.1 ) carrying three trait genes (neomycin phosphotransferase II [nptII], Cry1C and phosphinothricin acetyltransferase [Bar]). The latter two traits were transformed into the Oryza sativa indica cultivar Minghui 63 by Agrobacterium-mediated transformation20. T1c-19 exhibits high resistance to leaffolders (Cnaphalocrocis medinalis) and yellow stemborer (Tryporyza incertulas). The transgenic rice TT51-1 was co-transformed with two plasmids, pFHBT1 ( Supplemental sequence No.2 ) and pGL2RC7 ( Supplemental sequence No.3 ), into Minghui 63 by particle bombardment. These plasmids carry the trait genes Cry1Ab(c), bla and hpt (pFHBT1) and rice chitinase 2 (Cht-2) and hpt (pGL2RC7), respectively. The bla and hpt genes are not transcriptionally active in plants. Transformation resulted in insertion of the Cry1Ab(c), bla and hpt trait genes and thus in high resistance against yellow stem borers and leaf-folders18. Both transgenic rice events and their isogenic control lines were developed and supplied by Huazhong Agricultural University, China. The GM rice samples were subjected to whole genome sequencing and further analysis of PCR amplification. The isogenic non-GM rice line (Minghui 63) was used as a control to validate the results from bioinformatics analysis. Homogeneous seeds of transgenic rice T1c-19 and TT51-1 were planted in Wuhan (Hubei province, China) and harvested in September 2010.
Paired-end sequencing
Total genomic DNA was extracted from rice seeds using the DNeasy Plant Mini Kit (Qiagen). The quality of the extracted rice genomic DNA was evaluated by UV-spectrophotometry with a Nanodrop ND-8000 instrument (Thermo Scientific) and 1% agarose gel electrophoresis. The sequencing DNA libraries were created using Illumina Paired-End library preparation kits and Illumina Oligos for adaptor ligation according to the manufacturer's instructions (Illumina). The mean fragment size was about 500 bp. The constructed DNA libraries were quantified by Pico-Green (Quant-iT; Invitrogen) and Agilent Bioanalyzer DNA 1000 kit (Agilent Technologies). For both libraries 90 cycles of paired-end sequencing were performed on a single lane of an Illumina HiSeq 2000 DNA sequencer (Illumina). Paired-end sequencing and simple modification of the raw data to ensure sequencing quality were performed by BGI-Shenzhen (Shenzhen, China). Filtering was applied to remove reads of insufficient length, low read quality and to trim away adapter sequences before data analysis.
In silico mimicking of unknown insertion event
To test the potential for detection of unknown inserts (no a priori knowledge of transformation vector or insert), a set of paired-end reads was generated in silico and spiked into the raw data obtained with T1c-19 rice. First, a hypothetical unknown insertion event was designed by combining a part of contig 2 from the soybean event DP-305423-1 (patent WO2008054747) with rearranged rice sucrose phosphate synthase gene (accession no. D45890). Contig 2 is mainly composed of soybean-derived elements. This hypothetical unknown event should mimic insertion and partial rearrangement of the insertion locus and insert. The detailed structure of the mimic is described in Supplemental sequence No.11 . The mimic has 6424 bp, with 1874 A, 1374 T, 1213 C and 1962 G. The simulated 90 bp paired-end reads were generated based on the mimic, corresponding to a sequencing depth of 23.8 × ( Supplemental Table S9 ). The base error rate of each read was set to meet the uniformly decreasing distribution for an error rate of 0.1% at the start of the read increasing to 4% at the end of the read and 5% probability to have a random DNA read, maximum 1 N allowed in a given read, the insertion size is 472 bp (mean distance between ends of read-pairs) with the standard deviation of 11 bp. The reference mimic was defined as to have a mutation rate of 0.001, the fraction of mutations that are indels was defined to 0.1, indel extension probability was set to 0.3 and the minimum length indel was set to 1 bp.
Bioinformatics analysis and analytical program
Mapping of reads to the host genome (TIGR 7.0, including Chr1 to Chr12, Chloroplast, Mitochondrion, ChrSy and ChrUn) or transformation vector (pBar-1c, pFHBT1 and pGL2RC7) was performed using the Burrows-Wheeler Aligner (BWA) algorithm (version 0.6.2)21. All the mapping processes were based on the single-end strategy. In module 1, parameter -n (i.e. maximum edit distance) was set to 10, 20 and 30 while keeping the other parameters default. In module 2 and 3, we used the default mapping parameters in BWA. The Assembly by Short Sequences (ABySS) algorithm version 1.3.4 22 was used in all assembly processes, parameter k (i.e. size of k-mer) was set to 23 bp and the other parameters were default. The analytical pipeline of module 1 to module 3 analyses is implemented in Bash script under the Ubuntu X86_64 system. The source code and its executable file are available from the URL: http://gmdd.shgmo.org/Computational-Biology/Transeq/Transeq.tar.bz2).
Transgene element sequence library
A collection of altogether 134 sequence elements, mainly derived from transgenic plants and transformation vectors, were compiled from NCBI Genbank and GMDD databases25 and served as a transgene element sequence library ( Supplemental Table S3 ).
Experimental validation of transgenic inserts
The transgene location, flanking region sequences and the inserts of transgenic rice T1c-19 and TT51-1 were confirmed with conventional PCR and DNA sequencing. PCR amplifications were performed with the primers listed in Supplemental Table S2 and the PCR Amplification Kit (Takara Biotechnology Co., Ltd.) with the following cycling profile: 5 min at 94°C, 35 cycles of (30 sec denaturation at 94°C, 30 sec annealing at 52°C and 1 min extension at 72°C), followed by a 7 min additional extension step at 72°C. PCR products were separated by 2% agarose gel electrophoresis to visually inspect amplification products. Each product was purified from the gel using AxyPrep™ DNA Gel Extraction Kit (Axygen Biosciences) and successively cycle sequenced using BigDye Terminators Version 3.1 kit (Applied Biosystems) and the relevant primers ( Supplemental Table S2 ) on an ABI 3730 sequencer (Invitrogen).
Verification of the number of inserts
Independent verification of the number of inserts based on module 1 data was obtained with the equation (Equation 1):  , where X = insert detection index, N = number of reads mapped to the plasmid vector, Lr = read length, Lv1 = length of insert no.1, Lv2 = length of insert no.2, etc., D = sequencing depth and R = relative ratio of reads mapped to the host genome (detailed calculations described in
Table 1
). Theoretically, X = 1 if all inserts are detected and the sequencing data and analyses are perfect.
, where X = insert detection index, N = number of reads mapped to the plasmid vector, Lr = read length, Lv1 = length of insert no.1, Lv2 = length of insert no.2, etc., D = sequencing depth and R = relative ratio of reads mapped to the host genome (detailed calculations described in
Table 1
). Theoretically, X = 1 if all inserts are detected and the sequencing data and analyses are perfect.
References
Secretariat of the Convention on Biological Diversity. Secretariat of the Convention of Biological Diversity (2000).
Codex Alimentarius. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants. CAC/GL 45, 1–18 (2003).
Sparrow, P. GM risk assessment. Mol biotechnol 44, 267–275 (2010).
Miraglia, M. et al. Detection and traceability of genetically modified organisms in the food production chain. Food Chem Toxicol 42, 1157–1180 (2004).
Padgette, S. R. et al. Development, identification and characterization of a glyphosate-tolerant soybean line. Crop Sci 35, 1451–1461 (1995).
Astwood, J. D., Hammond, B. G., Dobert, R. C. & Fuchs, R. L. Updated molecular characterization and safety assessment of Roundup Ready soybean event 40-3-2. Monsanto Technical Report MSL-16712. St. Louis, Missouri. USA (2000).
Lirette, R. et al. Further molecular characterization of Roundup Ready soybean event 40-3-2. Monsanto Technical report MSL-16646. St. Louis, Missouri. USA (2000).
Windels, P., Taverniers, I., Depicker, A., Van Bockstaele, E. & De Loose, M. Characterisation of the Roundup Ready soybean insert. Eur Food Res Technol 213, 107–112 (2001).
Holst-Jensen, A. et al. Detecting un-authorized genetically modified organisms (GMOs) and derived materials. Biotechnol. Adv 30, 1318–1335 (2012).
Ruttink, T. et al. Molecular toolbox for the identification of unknown genetically modified organisms. Anal Bioanal Chem 396, 2073–2089 (2010).
Thudi, M., Li, Y., Jackson, S. A., May, G. D. & Varshney, R. K. Current state-of-art of sequencing technologies for plant genomics research. Brief Funct Genomics 11, 3–11 (2012).
Campbell, P. J. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet 40, 722–729 (2008).
Fullwood, M. J., Wei, C.-L., Liu, E. T. & Ruan, Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 19, 521–532 (2009).
Hormozdiari, F., Hajirasouliha, I., McPherson, A., Eichler, E. E. & Sahinalp, S. C. Simultaneous structural variation discovery among multiple paired-end sequenced genomes. Genome Res 21, 2203–2212 (2011).
Kovalic, D. et al. The Use of Next Generation Sequencing and Junction Sequence Analysis Bioinformatics to Achieve Molecular Characterization of Crops Improved Through Modern Biotechnology. The Plant Genome 5, 149–163 (2012).
DuBose, A. J. et al. Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene. Nucleic. Acids. Res. 41, e70 (2013).
Matsumoto, T. et al. The map-based sequence of the rice genome. Nature 436, 793–800 (2005).
Tu, J. et al. Field performance of transgenic elite commercial hybrid rice expressing Bacillus thuringiensis δ-endotoxin. Nat Biotechnol 18, 1101–1104 (2000).
Lu, C. The first approved transgenic rice in China. GM crops 1, 113–115 (2010).
Tang, W. et al. Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol Breeding 18, 1–10 (2006).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Simpson, J. T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res 19, 1117–1123 (2009).
Wu, G. et al. Real-time PCR method for detection of the transgenic rice event TT51-1. Food Chem 119, 417–422 (2010).
Latham, J. R., Wilson, A. K. & Steinbrecher, R. A. The mutational consequences of plant transformation. BioMed Research International, 25376 (2006).
Dong, W. et al. GMDD: a database of GMO detection methods. BMC Bioinformatics 9, 260 (2008).
Acknowledgements
This work was supported by the National Transgenic Plant Special Fund (2013ZX08012-002), the Shanghai Rising-Star Program (11QA1403300) and a grant from the Norwegian Research Council (178288) and tDECATHLON.
Author information
Authors and Affiliations
Contributions
L.T.Y. and C.M.W. did the data analysis, experimental validation and manuscript organizing; A.H. contributed to study design, data discussion and manuscript organizing and revision; D.M. contributed to data analysis, discussion and manuscript revision; Y.J.L. developed and supplied the transgenic rice materials; D.B.Z. conceived and coordinated this work. All authors read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplemental Figures 1-8 and Sequences 1-13
Supplementary Information
Supplemental Tables S1-S9
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yang, L., Wang, C., Holst-Jensen, A. et al. Characterization of GM events by insert knowledge adapted re-sequencing approaches. Sci Rep 3, 2839 (2013). https://doi.org/10.1038/srep02839
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02839
This article is cited by
-
TC-hunter: identification of the insertion site of a transgenic gene within the host genome
BMC Genomics (2022)
-
A paired-end whole-genome sequencing approach enables comprehensive characterization of transgene integration in rice
Communications Biology (2022)
-
Development and evaluation of triple gene transgenic cotton lines expressing three genes (Cry1Ac-Cry2Ab-EPSPS) for lepidopteran insect pests and herbicide tolerance
Scientific Reports (2022)
-
Whole-genome resequencing using next-generation and Nanopore sequencing for molecular characterization of T-DNA integration in transgenic poplar 741
BMC Genomics (2021)
-
Influence of transgenesis on genome variability in cucumber lines with a thaumatin II gene
Physiology and Molecular Biology of Plants (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.