Abstract
To extend agricultural productivity by knowledge-based breeding and tailor varieties adapted to specific environmental conditions, it is imperative to improve our ability to assess the dynamic changes of the phenome of crops under field conditions. To this end, we have developed a precision phenotyping platform that combines various sensors for a non-invasive, high-throughput and high-dimensional phenotyping of small grain cereals. This platform yielded high prediction accuracies and heritabilities for biomass of triticale. Genetic variation for biomass accumulation was dissected with 647 doubled haploid lines derived from four families. Employing a genome-wide association mapping approach, two major quantitative trait loci (QTL) for biomass were identified and the genetic architecture of biomass accumulation was found to be characterized by dynamic temporal patterns. Our findings highlight the potential of precision phenotyping to assess the dynamic genetics of complex traits, especially those not amenable to traditional phenotyping.
Similar content being viewed by others
Introduction
Plant breeding has substantially contributed to the increase in agricultural productivity during the last century1 but satisfying the needs of a growing human population still presents a tremendous challenge for crop improvement2. While recently developed genomic approaches promise to dramatically increase progress by breeding3, our ability to characterise the phenome of a plant in the field has changed little since the advent of science-based plant breeding more than 100 years ago. This phenotyping bottleneck is of particular severity since many traits of biological and agricultural importance are under the control of complex dynamic regulation4. For example, biomass changes with plant development over time but traditional approaches to unravel the genetic architecture underlying such traits focused on single time points thus neglecting the developmental dynamics of trait formation. A key component to maintain or even increase agricultural production is, therefore, the development of phenotyping technologies that enable monitoring the phenotypic changes of crop plants in the field5,6.
The accumulation of biomass is central to agricultural productivity but the non-invasive monitoring of biomass using various sensors has thus far yielded only moderate prediction accuracies7,8,9. This may be attributed to the failure in combining information derived from different but complementary types of sensors10. Triticale (x Triticosecale Wittmack L.; AABBRR; 2n = 6x = 42), the interspecific cross between wheat and rye, is mainly used as animal feed but also shows great potential as bioenergy crop and is thus well suited to study the dynamics of biomass accumulation in small grain cereals. Here, we present a novel precision phenotyping platform for non-invasive high-throughput phenotyping of small grain cereals under field conditions and its application to dissect the genetic architecture of biomass accumulation by a genome-wide association study.
Results
Precision phenotyping platform
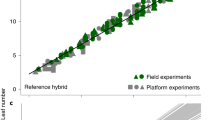
We have developed a novel precision phenotyping platform that enables high-throughput and high-dimensional phenotyping of small grain cereals in the field (Fig. 1). Our precision phenotyping platform permits the collection of data from complementary types of sensors and incorporates light curtains, laser distance sensors, 3D-Time-of-Flight cameras and high-resolution hyperspectral imaging11. In addition, it allows for sensor fusion to accurately predict biomass under field conditions (Fig. 1b,c). We conducted a detailed calibration experiment for our precision phenotyping platform based on 1,200 biomass yield plots of triticale. The technical repeatability of the different single sensor measurements was high (Table S1). The platform enables high-throughput screening of more than 2,000 plots per day which in combination with the nearly fully automated subsequent data analysis facilitates rapid and economic trait determination. The technical repeatability of the sensor fusion calibration model for biomass was very high (Fig. 1d) and we obtained excellent prediction accuracies for biomass at all three developmental stages (Fig. 1e). Transferability of the calibration models across environments was evaluated by predicting biomass yields of the year 2012 based on the data from 2011 and vice versa. This yielded high cohort validated R2 values of 0.93 and 0.84 (Fig. S3).
Precision phenotyping platform.
(a,b) Platform with multiple sensors for non-invasive assessment of biomass under field conditions. 3D-ToF: 3D-Time-of-Flight camera; LDS: laser distance sensor; HSI: hyperspectral imaging; LCI: light curtain imaging. (c) Information captured by the different sensors in a single yield plot. (d) Technical repeatability and (e) prediction accuracy of the platform based on sensor fusion models using data from two years.  and
and  denote the coefficient of determination of cross-validation and of repetition, respectively and RMSREv and RMSREw denote the root mean squared relative error of cross-validation and of repetition, respectively.
denote the coefficient of determination of cross-validation and of repetition, respectively and RMSREv and RMSREw denote the root mean squared relative error of cross-validation and of repetition, respectively.
Temporal genetic patterns of biomass accumulation
Using the precision phenotyping platform and the developed calibration models we predicted biomass at three developmental stages (BM1–BM3) in a large mapping population of 647 doubled haploid triticale lines (Fig. 2a, S4a). The predicted biomass data were highly heritable with heritabilities ranging from 0.78 to 0.84 (Table 1, S3). We used a genome-wide association mapping scan to identify QTL associated with biomass at each of the three developmental stages (Table 1, Fig. 2, S5, S6a). A detailed analysis of the linkage disequilibrium structure in the genomic regions associated with biomass (Fig. S7–S10) and the collinearity among markers (Table S4) revealed two major QTL regions on chromosomes 5A and 5R with effects on biomass (Fig. S11). The markers most closely linked to the two major QTL and thus candidates for a marker-assisted selection are wPt-2329 on chromosome 5A and rPt-509721 and rPt-399681 on chromosome 5R. Together, all identified QTL explained 40.14, 31.55 and 28.52% of the genetic variance of predicted BM1, BM2 and BM3, respectively.
Genetic architecture of biomass accumulation.
(a) Schematic representation of small grain cereal growth and the three developmental stages at which biomass (BM) was assessed in this study. (b) Venn diagram for markers significantly associated with BM1, BM2, BM3 and in the multivariate analysis. (c) Manhattan plots of the genome-wide association study. Significant associations are shown in green.
A multivariate analysis incorporating BM1, BM2 and BM3, jointly identified the same two major QTL on chromosomes 5A and 5R that have been identified in the analyses of the single developmental stages (Fig. 2, S4b, S6b, Table S5, S6). In addition, growth curve parameters were estimated for each line and used for QTL mapping (Fig. S13, Table S7, S8). To dissect the genetic architecture underlying biomass accumulation, we performed full 2-dimensional epistasis scans. Epistatic QTL for biomass were detected for all three developmental stages (Fig. 3).
Discussion
Genomic approaches to dissect the genetic architecture of complex traits rely on both high-quality genotypic and phenotypic data. While the power of current genotyping technologies enables high-density genotyping or even resequencing of entire genomes, our ability to measure traits in the field is limited. Advances in sensor technologies, however, now offer an array of tools that can be deployed to overcome this phenotyping bottleneck and bring a revolution to the assessment of the plant phenome. The precision phenotyping platform presented here combines various sensors for a non-invasive assessment of small grain cereals under field conditions11. With a capacity of more than 2,000 plots per day for data collection and post-processing of the collected raw data, it outperforms the phenotyping capacity of a person and, most importantly, permits the collection of data for novel traits not amenable to traditional phenotyping (e.g., biomass, stress tolerance, or primary and secondary metabolites).
The limited prediction accuracies of the calibration models based on single sensors (Table S1) clearly emphasized the need to integrate data from multiple sensors to capture complementary information on plant characteristics (Table S2). This is, for example, illustrated by the improvement in biomass prediction accuracy by considering dry matter content which can accurately be predicted with hyperspectral imaging (Fig. S1, S2). The high technical repeatability (Fig. 1d), the high prediction accuracies (Fig. 1e) and the high heritability (Table 1) illustrate that the sensors and the sensor fusion calibration models as employed here are well suited for the non-invasive assessment of biomass yield. A parameter of eminent relevance for plant breeding is the transferability of the established calibration models across environments. The high accuracies obtained for calibration models established in one year and predicting in the other year demonstrate the robustness of the approach (Fig. S3). In conclusion, the precision phenotyping platform facilitates the non-invasive collection of high-quality, multi-dimensional and high-throughput phenotypic data under field conditions.
A large mapping population of 647 doubled haploid triticale lines derived from four crosses was employed to determine the heritable portion of variation for biomass accumulation and dissect the underlying genetic architecture. The biomass data predicted based on the developed calibration models were highly heritable at all three developmental stages (Table 1, S3). These high heritabilities in combination with the high prediction accuracies illustrate the great potential of the precision phenotyping platform for genomics approaches. Genome-wide association mapping scans at the three developmental stages identified two major QTL regions associated with biomass yield (Fig. 2). The moderate predictive power for the proportion of explained genotypic variance obtained with the identified QTL (Table 1) suggests that biomass must be regarded as highly complex trait with many small effect QTL12 that escape detection in QTL mapping approaches. Consistent with the complex nature of the trait, grain yield, heading time, spikes per square meter, 1000-kernel weight and early plant height have recently been identified as key contributors to early biomass13. Our results illustrate the great importance of non-invasively assessing different time points in plant development as a prerequisite for knowledge-based adaptation breeding to tailor cultivars to local climatic conditions (Fig. S12).
Our analysis of the main effect QTL for biomass at the three developmental stages revealed a dynamic genetic pattern underlying biomass accumulation (Fig. 2). Whereas the major QTL on chromosome 5R is active throughout plant development, the other major QTL on chromosome 5A shows a temporal pattern of activity. It contributes strongly to biomass at the early stage, then its activity ceases and by the last developmental stage it has completely discontinued its contribution to biomass accumulation. This may also be caused by the temporal contribution of one of the above-mentioned traits with a causal relation to biomass, for example heading time. The observed temporal activity of QTL motivated us to perform a QTL scan accommodating the multivariate nature of the developing trait which confirmed the need for a temporal assessment because many QTL would remain undetected by the traditional static examination of only a single time point in development (Fig. 2, S4b, S6b, Table S5, S6). The growth of organisms has recently been shown to follow a sigmoid curve based on fundamental principles for the allocation of metabolic energy between maintenance of existing tissue and the production of new biomass14. As an alternative approach we therefore determined growth curve parameters based on a logistic growth function, a common sigmoid function and used these for QTL mapping4. This approach confirmed that biomass accumulation is to a large extent genetically determined (Table S7) and the identified QTL further substantiate the dynamic genetics underlying the trait (Fig. S13, Table S8). In conclusion, a temporal assessment of the phenome, as facilitated by the developed precision phenotyping platform, is of paramount importance to unravel the complex genetic patterns underlying the expression of dynamic traits. This knowledge can in turn assist the selection of lines with complementary pattern types as crossing parents.
The observed temporal activity of individual QTL suggested that also interactions among loci may show patterns of genetic regulation during plant development. Epistasis refers to the dependency of the effect of an allele at one genetic locus on the allele status at one or multiple other loci15. Recent work at the level of proteins has provided evidence that epistasis is as key player regulating evolution and potentially also fitness levels of individuals within a population16. In accordance with this, epistasis has been shown to contribute to the genetic architecture of complex traits in crops17,18. Employing full 2-dimensional epistasis scans we detected epistatic QTL for biomass at all three developmental stages (Fig. 3). The proportion of explained genotypic variance by single epistatic QTL was small, but given their high number we speculate that epistasis contributes substantially to the heritability of biomass in small grain cereals. Our results on the complex trait biomass thus corroborate those from a recent theoretical approach showing that the contribution of epistasis increases with the biological complexity of the trait19. Interestingly, we found that the main effect QTL detected on chromosome 5R is also involved in a high number of epistatic interactions with loci throughout the genome. Such epistatic master regulators have recently been described in Arabidopsis, wheat and sugar beet20,21,22. A possible molecular mechanism mediating such associations has been provided by the discovery that the human transcription factor KLF14 acts as trans master regulator of adipose gene expression23 suggesting that transcriptional regulation may in part explain the observed epistatic interactions. As for the main effect QTL, we observed temporal patterns also for the epistatic interactions. Our results thus illustrate that the entire genetic architecture of biomass accumulation in triticale is under dynamic control.
In a wider context, our discoveries on biomass accumulation in triticale are likely to have broad relevance to other crops as well as other traits showing dynamic genetic patterns of regulation. We anticipate that with the incorporation of information from additional sensors other agronomic important traits like for example disease resistances can be dynamically phenotyped. On the basis of our results we conclude that precision phenotyping platforms may become the method of choice to assess the genetics of complex dynamic traits under field conditions.
Methods
Plant material and field trials
For this study we used triticale as model species for small grain cereals. Two experiments were conducted: one for the calibration of the precision phenotyping platform and a second experiment aimed at dissecting the genetic architecture of biomass accumulation. The calibration experiment was based on 25 diverse triticale lines that were grown at one location (Stuttgart-Hohenheim, Germany) in two years under two plant densities (280 plants per m2 as optimum and reduced to 140) and two N fertilizer schemes per plant density (standard practice and 50% reduction), with two replicates per treatment combination. The plants were phenotyped with the precision phenotyping platform and subsequently harvested with a field chopper at approximately BBCH stage 49 (awns visible), 69 (late flowering) and 81 (very early dough development)24 to determine the reference fresh weight. A sample of every plot was dried to determine dry matter content and dry biomass yield.
The second experiment was based on a mapping population of 647 doubled haploid triticale lines25 derived from four families designated AxB (131), AxC (120), DxE (200) and DxF (196) which have been described by Alheit et al.26 as populations DH6, DH7, EAW74 and EAW78. The lines from the mapping population were grown in partially replicated designs27 including common checks with 960 plots per location at a plant density of 280 plants per m2. The mapping population was grown at two locations (Germany: Stuttgart-Hohenheim, 48.77° latitude, 9.18° longitude; Bohlingen, 47.72° latitude, 8.9° longitude) in two years (2011 and 2012). All plants were assessed with the phenotyping platform between 9 AM and 6 PM at approximately BBCH stage 49, 69 and 81 to predict biomass based on the calibration models established in the calibration experiment. The growth parameters (μ, λ, integral) were determined with the R package grofit28. The growth rate is expressed by the maximum slope μ, λ is the length of the lag phase and the integral corresponds to the area under the curve (Fig. S13).
Association mapping
The plants were genotyped with 1710 DArT markers and the map positions of a consensus map were used for the analysis26. Linkage disequilibrium (LD) was measured as r2 [ref 29] and calculated with software package Plabsoft30. Genome-wide association mapping was done with a mixed model approach incorporating kinship information21,31,32. For main effect QTL and for epistatic QTL, the Bonferroni-Holm procedure33 was applied to correct for multiple testing with P < 0.05. In addition, a multivariate mixed model approach with different models for the variance structure was used to allow for correlations among the three developmental stages. All mixed model calculations were performed using the software ASReml 3.034. The proportion of genotypic variance (pG) explained by the detected QTL was calculated by fitting all QTL simultaneously in the order of the strength of their association with the trait in a linear model including a family effect to obtain the sums of squares of the QTL (SSQTL). Thus, pG = SSQTL/h2, where h2 refers to the heritability32. The circular plots illustrating the epistatic interactions were created with Circos35. The results from the QTL mapping are available as supplementary data.
Phenotyping platform
We implemented a sensor platform using a tractor pulled trailer equipped with two light curtains (Infrascan 5000, Sitronic GmbH, Steyregg, Austria), three laser distance sensors (LDS1: OADM 96k/V66-2300-S12, Leuze Electronic GmbH + Co. KG, Owen, Germany; LDS2 and LDS3: OADM 20I6480/S14F, Baumer Holding AG, Frauenfeld, Switzerland), two 3D-Time-of-Flight cameras (Effector3D, Ifm Electronic GmbH, Essen, Germany) and a hyperspectral imaging system (Helios Core NIR, EVK DI Kerschhaggl GmbH, Raaba, Austria) equipped with a 120 W halogen lighting system (Figure 1). The sensors were mounted at the back of the trailer in a separate height adjustable sensor module which was shaded with a black canvas to avoid influence of direct solar radiation. A detailed technical overview of the developed precision phenotyping platform including descriptions of the mechanical design, the integrated sensor systems, the hard- and software design for plot based data collection and analysis and the phenotyping procedure are given in Busemeyer et al.11.
Biomass prediction
The model to determine the biomass of the plots is based on the fusion of parameters with selectivity to (i) the volume of the plants and (ii) their density, both extracted from sensor raw data for each plot. These parameters were fused and related to the dry biomass of each plot of the calibration experiment by multiple linear regression analysis to generate a calibrated biomass determination model.
The density of a plant is related to its dry matter content. Consequently, we used the hyperspectral imaging system to determine the dry matter content of the plants (which is physically based on the selectivity of the spectral imaging system to the plants' moisture content) and used this information as an approximation for the density of the plants. The automated analysis of spectral imaging data was performed with an application developed with the software package MATLAB (The Math Works, Natick, USA) including all steps of data transformation between sensor raw data and predicted dry matter content for each plot. In a first step noise occurring in the raw data coming from uneven spectral sensitivities of the sensor and a spatially unbalanced illumination was compensated by dividing all measured spectra by a reference. These reference spectra were generated under controlled conditions by placing a reference object made of Spectralon® in the sensor's field of view. In the next step the spectral values of a plot were split into two data sets corresponding to values from plants and soil based on the spectral angle mapper method36 with a threshold α of 0.03. Angles below this threshold were classified as spectra belonging to plants and the remaining spectra were excluded from further analysis. To reduce the influence of different reflection intensity levels due to shaded parts of the plants and different distances of the plants to the sensor, all spectra were normalized to a value of 1 at the wavelength of 1050 nm which is a wavelength nearly unaffected by the water content of the plants. Subsequently, all spectra belonging to plants were averaged to a single spectrum for each plot and the first derivative of the spectrum was calculated for a baseline shift correction. The pre-processed hyperspectral data of the plants were used in combination with a principle component analysis (PCA) using the MATLAB function plsregress to develop a calibration model for non-invasive determination of dry matter content of a plot.
We defined and extracted different parameters with selectivity to the volume of the plots from the raw data of the 3D-Time-of-Flight cameras, the light-curtains and the laser distance sensors. The automated data analysis for all different types of sensors was performed with an application developed with the software package MATLAB. The data of the 3D-Time-of-Flight cameras was used to estimate the following parameters for each plot: (i) “Plant height” calculated as the difference between the average of the 1% maximum and 1% minimum distances. (ii) “Penetration-depth-top-3D” calculated as the mean value of all distance values minus the average value of the 1% minimum distances. (iii) “Penetration-depth-sidewise-3D” estimated as the mean value of all distance values measuring from side view into the plants.
The data of the light-curtains was used to extract the following parameters for each plot: (i) “Plant height” was determined as the average value of the 1% highest interrupted light barriers in combination with the laser distance sensor mounted at the bottom edge of the light-curtain which delivers the distance of the light-curtains to the ground. (ii) “Coverage density” was estimated as the percentage of interrupted light barriers of the lower light-curtain of a plot. To avoid a possible impairment of plots with low plant heights, the algorithm first determines the canopy of the plot and then only takes into account the light barriers underneath the average top edge of the plants.
As a third approach, we used data of the laser distance sensors and estimated the following parameters for each plot: (i) “Plant height” was measured with LDS1 as the difference between the average value of the 3% maximum and 3% minimum distances. (ii) “Penetration-depth-top” was estimated with LDS1 as the mean value of all distance values minus the average value of the 3% minimum distances. (iii) “Penetration-depth-sidewise” was determined with LDS2 as the mean value of all distance values. (iv) “Leap-rate” was calculated based on data of LDS2. Distance leaps between consecutive raw data values >2 cm were interpreted as intersections between two single plants. ”Leap-rate” refers to the sum of detected distance leaps and was used as an approximation for the number of plants.
The different parameters with selectivity to the volume and the average density of the plots and plants, respectively, were fused and related to plant biomass with multiple linear regression to generate a biomass prediction model:

where  denotes the observed biomass for each plot in the field,
denotes the observed biomass for each plot in the field,  the number of parameters extracted out of one class of sensors but also across sensors,
the number of parameters extracted out of one class of sensors but also across sensors,  the ith parameter of the model,
the ith parameter of the model,  the regression coefficients and
the regression coefficients and  the error term. The logarithmic model was chosen, because approximation of volume is based on a multiplicative action of single parameters. Regression analysis was performed with function mvregress of software package MATLAB. We computed maximum likelihood estimates with a limit of 100 iterations and the default settings of convergence tolerance for changes of beta and the objective function. We applied a forward model selection algorithm with function stepwisefit of software package MATLAB with an entrance tolerance of P < 0.01 and an exit tolerance of P < 0.05 to assess the contribution of the different parameters to the described variance in the model and to select a final sensor fusion calibration model of biomass.
the error term. The logarithmic model was chosen, because approximation of volume is based on a multiplicative action of single parameters. Regression analysis was performed with function mvregress of software package MATLAB. We computed maximum likelihood estimates with a limit of 100 iterations and the default settings of convergence tolerance for changes of beta and the objective function. We applied a forward model selection algorithm with function stepwisefit of software package MATLAB with an entrance tolerance of P < 0.01 and an exit tolerance of P < 0.05 to assess the contribution of the different parameters to the described variance in the model and to select a final sensor fusion calibration model of biomass.
Evaluation of the prediction models
We determined the quality of our calibrations estimating (i) the root mean squared relative error of calibration as a measure for the accuracy of the biomass predictions as

where  denotes the number of samples in the dataset,
denotes the number of samples in the dataset,  the predicted and
the predicted and  the observed biomass of the ith plot and (ii) the coefficient of determination of calibration between observed and predicted biomass
the observed biomass of the ith plot and (ii) the coefficient of determination of calibration between observed and predicted biomass  . Moreover, over-fitting of the model was tested by applying 5-fold cross validation. Cross validation was repeated 100 times and the root mean squared relative error (RMSREv) and coefficient of determination (
. Moreover, over-fitting of the model was tested by applying 5-fold cross validation. Cross validation was repeated 100 times and the root mean squared relative error (RMSREv) and coefficient of determination ( ) of validation were averaged across runs.
) of validation were averaged across runs.
Transferability of the calibrations across environments was determined applying cohort validation. Biomass prediction models were calibrated with data collected in the year 2011 and predicted plot dry biomass yield of year 2012 and vice versa. For the cohort validations we estimated the bias μ between the predicted and the observed values as

where  denotes the number of samples in the dataset,
denotes the number of samples in the dataset,  the predicted and
the predicted and  the observed biomass of the ith plot. This bias indicates a general over- or underestimation of biomass yield. Furthermore, the root mean squared relative error (RMSREcv) and the coefficient of determination were determined for the cohort validation (
the observed biomass of the ith plot. This bias indicates a general over- or underestimation of biomass yield. Furthermore, the root mean squared relative error (RMSREcv) and the coefficient of determination were determined for the cohort validation ( ).
).
Technical repeatability
To assess the precision of the developed precision phenotyping platform, every plot was recorded twice within a repetition time of less than 10 minutes except for BM3 in year 2011 where the repetition time was about 90 minutes. The technical repeatability was determined by comparing the two measurements with linear regression. To quantify the results, the following statistics were applied: (i) root mean squared relative error of repetition

where m denotes the number of repeated samples in the dataset,  the first and
the first and  the second repetition of the same plot and (ii) the coefficient of determination of repetition (
the second repetition of the same plot and (ii) the coefficient of determination of repetition ( ).
).
References
Evenson, R. E. & Golin, D. Assessing the impact of the green revolution, 1960 to 2000. Science 300, 758–762 (2003).
Takeda, S. & Matsuoka, M. Genetic approaches to crop improvement: responding to environmental and population changes. Nature Rev. Genet. 9, 444–457 (2008).
Edwards, D., Batley, J. & Snowdon, R. J. Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 126, 1–11 (2013).
Wu, R. & Lin, M. Functional mapping – how to map and study the genetic architecture of dynamic camplex traits. Nature Rev. Genet. 7, 229–237 (2006).
White, J. W. et al. Field-based phenomics for plant genetics research. Field Crops Res. 133, 101–112 (2012).
Montes, J. M., Melchinger, A. E. & Reif, J. R. Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci. 12, 433–436 (2007).
Ehlert, D., Horn, H.-J. & Adamek, R. Measuring crop biomass density by laser triangulation. Comput. Electron. Agric. 61, 117–125 (2008).
Ehlert, D., Heisig, M. & Adamek, R. Suitability of a laser rangefinder to characterize winter wheat. Precision Agric. 11, 650–663 (2010).
Erdle, K., Mistele, B. & Schmidhalter, U. Comparison of active and passive spectral sensors in discriminating biomass parameters and nitrogen status in wheat cultivars. Field Crop Res. 124, 74–84 (2011).
Ruckelshausen, A. Autonomous robots in agricultural field trials. In: Bleiholder H., Piepho H.-P. (Eds.), Proceedings of the International Symposium “Agricultural Field Experiments – Today and Tomorrow”, 190–197 (2007).
Busemeyer, L. et al. BreedVision – A multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors 13, 2830–2847 (2013).
Mackay, T. F. C., Stone, E. A. & Ayroles, J. F. The genetics of quantitative traits: challenges and prospects. Nature Rev. Genet. 10, 565–577 (2009).
Gowda, M. et al. Potential for simultaneous improvement of grain and biomass yield in Central European winter triticale germplasm. Field Crops Res. 121, 153–157 (2011).
West, G. B., Brown, J. H. & Enquist, B. J. A general model for ontogenetic growth. Nature 413, 628–631 (2001).
Carlborg, Ö. & Haley, C. S. Epistasis: too often neglected in complex trait studies? Nature Rev. Genet. 5, 618–625 (2004).
Breen, M. S., Kemena, C., Vlasov, P. K., Notredame, C. & Kondrashov, F. A. Epistasis as the primary factor in molecular evolution. Nature 490, 535–538 (2012).
Buckler, E. S. et al. The genetic architecture of maize flowering time. Science 325, 714–718 (2009).
Würschum, T., Maurer, H.-P., Dreyer, F. & Reif, J. C. Effect of inter- and intragenic epistasis on the heritability of oil content in rapeseed (Brassica napus L.). Theor. Appl. Genet. 126, 435–441 (2013).
Zuk, O., Hechter, E., Sunyaev, S. R. & Lander, E. S. The mystery of missing heritability: genetic interactions create phantom heritability. PNAS 109, 1193–1198 (2012).
Wentzell, A. M., Boeye, I., Zhang, Z. & Kliebenstein, D. J. Genetic networks controlling structural outcome of glucosinolate activation across development. PloS Genet. 4, e1000234 (2008).
Reif, J. C. et al. Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theor. Appl. Genet. 123, 283–292 (2011).
Würschum, T. et al. Genome-wide association mapping of agronomic traits in sugar beet. Theor. Appl. Genet. 123, 1121–1131 (2011).
Small, K. S. et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nature Genet. 43, 561–564 (2011).
Lancashire, P. D. et al. A uniform decimal code for growth stages of crops and weeds. Ann. Applied Biology 119, 561–601 (1991).
Würschum, T., Tucker, M. R., Reif, J. C. & Maurer, H.-P. Improved efficiency of doubled haploid generation in hexaploid triticale by in vitro chromosome doubling. BMC Plant Biology 12, 109 (2012).
Alheit, K. V. et al. Detection of segregation distortion loci in triticale (x Triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics 12, 380 (2011).
Williams, E., Piepho, H.-P. & Whitaker, D. Augmented p-rep designs. Biometrical Journal 53, 19–27 (2011).
Kahm, M. et al. grofit: Fitting biological growth curves with R. Journal of Statistical Software Vol. 33, Issue 7 (2010).
Weir, B. S. Genetic data analysis II. 2nd edn. Sinauer Associates, Sunderland (1996).
Maurer, H. P., Melchinger, A. E. & Frisch, M. Population genetic simulation and data analysis with Plabsoft. Euphytica 161, 133–139 (2008).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genet. 38, 203–208 (2006).
Würschum, T. et al. Comparison of biometrical models for joint linkage association mapping. Heredity 108, 332–340 (2012).
Holm, S. A Simple Sequentially Rejective Bonferroni Test Procedure. Scand. J. Stat. 6, 65–70 (1979).
Gilmour, A. R., Gogel, B. J., Cullis, B. R. & Thompson, R. ASReml User Guide, Release 3.0. VSN International Ltd, Hemel Hempstead, HP1 1ES, UK (2009).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645 (2010).
Park, B., Windham, W. R., Lawrence, K. C. & Smith, D. P. Contaminant classification of poultry hyperspectral imagery using spectral angle mapper algorithm. Biosystems Engineering 96, 323–333 (2007).
Acknowledgements
This research was funded by the German Federal Ministry of Education and Research (BMBF) under the promotional reference 0315414.
Author information
Authors and Affiliations
Contributions
J.C.R., A.R., T.W., H.P.M., V.H. designed the study. L.B., K.M., A.R. constructed the phenotyping platform. L.B., K.M., K.V.A., H.P.M., E.A.W. collected phenotypes. L.B. and T.W. carried out analyses. T.W., L.B., A.E.M., J.C.R. wrote the paper. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
QTL mapping data
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Busemeyer, L., Ruckelshausen, A., Möller, K. et al. Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci Rep 3, 2442 (2013). https://doi.org/10.1038/srep02442
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02442
This article is cited by
-
Early prediction of biomass in hybrid rye based on hyperspectral data surpasses genomic predictability in less-related breeding material
Theoretical and Applied Genetics (2021)
-
Scaling up high-throughput phenotyping for abiotic stress selection in the field
Theoretical and Applied Genetics (2021)
-
Identification of quantitative trait loci associated with canopy temperature in soybean
Scientific Reports (2020)
-
Accelerating crop genetic gains with genomic selection
Theoretical and Applied Genetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.