Abstract
Despite significant advancements with recombinant AAV2 or AAV8 vectors for liver directed gene therapy in humans, it is well-recognized that host and vector-related immune challenges need to be overcome for long-term gene transfer. To overcome these limitations, alternate AAV serotypes (1–10) are being rigorously evaluated. AAV5 is the most divergent (55% similarity vs. other serotypes) and like AAV1 vector is known to transduce liver efficiently. AAV1 and AAV5 vectors are also immunologically distinct by virtue of their low seroprevalence and minimal cross reactivity against pre-existing AAV2 neutralizing antibodies. Here, we demonstrate that targeted bio-engineering of these vectors, augment their gene expression in murine hepatocytes in vivo (up to 16-fold). These studies demonstrate the feasibility of the use of these novel AAV1 and AAV5 vectors for potential gene therapy of diseases like hemophilia.
Similar content being viewed by others
Introduction
Recombinant adeno-associated virus (AAV) vectors have shown significant promise for gene therapy, particularly when targeted to post-mitotic and/or immune-privileged tissues1,2,3,4,5. Clinical trials with the prototype AAV2 vectors in patients with hemophilia B, established the safety of the AAV based vector system for muscle- or liver-directed gene transfer6,7. However, insufficient transgene expression and immune reactions directed against the AAV2 capsid precludes its widespread use8,9. As a consequence, strategies such as the use of alternate serotypes (AAV1-10)10,11, hybrid/shuffled AAV vectors12 and transient immuno-suppression protocols13,14 are being explored to overcome some of these limitations. Of these, AAV8 serotype vector has demonstrated sustained therapeutic levels of factor (F) IX in a liver directed clinical trial in patients with hemophilia B15. However, in this trial, two patients who received the highest dose (2 × 1012 viral genome (vg)/kg) of the vector developed capsid specific T cells that required glucocorticoid therapy to attenuate this response. Thus, the theme of AAV vector dose dependent immunotoxicity seen earlier with AAV2 vectors seems to re-emerge with AAV8 vectors as well. In this context, recent studies that have generated modified AAV2 or AAV8 vectors that provide enhanced gene expression at significantly lesser vector-dose becomes significant16,17,18.
Despite the availability of these improved AAV2 and AAV8 vectors, it is important to note that human beings are natural hosts for AAV19,20. Primary infection often generates anti-AAV neutralizing antibodies (NAb), which are prevalent in the general population19. The degree of seroprevalance against AAV varies in different populations19,20. Available data suggests that the highest incidence of pre-existing NAb is against AAV2 vectors (upto ~43.5–72%), while the lowest (~22–38%) is against AAV8 vectors19,21. This is particularly important since even low levels of NAb are known to completely abrogate transduction with high titers of vectors administered22,23. In addition, NAb specific for AAV2 are known to cross-neutralize AAV8 to certain extent21. The presence of and the variation in such pre-existing immunity against different AAV serotypes in humans suggests that no one AAV serotype will be universally applicable for therapeutic gene transfer. Thus it becomes important to utilize and develop other alternate AAV serotypes which can transduce the liver to have widespread clinical applications. AAV1 has been reported to efficiently transduce muscle24,25, while AAV5 is efficient in transducing lungs and muscle tissue26,27. Both AAV1 and AAV5 are also known to be efficient in liver directed gene transfer in pre-clinical models10,23,28 and have low cross-reactivity with AAV2 neutralizing antibodies24,27,29. Indeed, pseudotyped AAV1 and AAV5 vectors are already in Phase I/II clinical trials for treating alpha-1 antitrypsin deficiency (ClinicalTrials.gov identifier- NCT00430768), muscular dystrophy (ClinicalTrials.gov identifier- NCT01344798) and lipoprotein lipase deficiency30.
We have recently developed novel AAV2 and AAV8 vectors by targeted modification of capsid phosphodegrons which circumvents the host-cellular serine/threonine kinase phosphorylation and ubiquitination of the viral capsid16,18. These modifications improved the hepatic gene expression of AAV2 and AAV8 vectors by up to 14 and 46-fold, respectively. Since a majority of these residues are conserved in the VP1-3 capsid region of both AAV1 and AAV5 vectors and in anticipation of improving their transduction efficiency, we performed targeted mutagenesis in AAV1 and AAV5 vectors and evaluated them in vitro and in vivo.
Results
Selection of mutation targets in AAV1 and AAV5 capsid
We generated a total of seven each of AAV1 and AAV5 serine(S)/threonine(T) → alanine(A), lysine(K) → arginine(R) mutant vectors (Supplementary Figure S1 and S2). Four serine residues were selected for mutagenesis in AAV1 and AAV5 based on sequence comparison between the effective AAV2 capsid mutants (S276, S489, S498, S662, S668) generated earlier16. In case of threonine targets, the best performing AAV2 mutant T251A16, had a homolog residue in AAV1 (T251) but not in AAV5 capsid. Therefore two additional targets in AAV5 capsid (T107, T328) were modified based on their high conservation across AAV 1–10 (Supplementary Figure S2). The lysine residues in both AAV1 (K137) and AAV5 (K32) were chosen based on the highest probability of being ubiquitinated as predicted by the UbPred software (www.ubpred.org). A control mutation at a serine residue (S526A) in AAV1 that was conserved only in 5 out of 10 AAV serotypes was chosen as a negative control for the mutation experiments. When compared to the wild-type (WT) vectors, the S/T/K mutant capsids had comparable vector packaging efficiency (4 × 1011 vg/mL to 2 × 1012 vg/mL for AAV1 and 4 × 1011 vg/mL to 1.6 × 1012 vg/mL for AAV5) suggesting that modification of these specific amino acids had negligible effect on the capsid structure.
AAV5 serine and threonine mutant vectors improve gene transfer efficiency in vitro and in vivo
We first evaluated each of the S/T/K mutant AAV5 vectors expressing enhanced green fluorescent protein (EGFP) for their transduction efficiency in Chinese Hamster Ovary (CHO) cells at a multiplicity of infection (MOI) of 5000 vgs/cell. As can be seen in Figure 1a, all the AAV5 mutant vectors tested had superior transduction efficiency (79–97% vs. 50%) when compared to cells infected with WT-AAV5 vectors alone. The maximal increase was seen with AAV5-S652A vector (97%) followed by cells infected with S268A vectors (94%) (Figure 1a). Interestingly, in our previous studies the AAV5-S652A equivalent residue in AAV2 vector-S662A also demonstrated a similar increase (75% vs. 40% in WT-AAV2 infected cells) in transduction efficiency in vitro16.
AAV5 serine/threonine mutant vectors demonstrate increased transduction efficiency in vitro and in vivo.
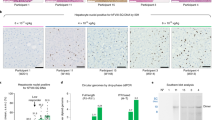
(a) In vitro transduction efficiency of AAV5 vectors. CHO cells were either mock-infected or infected with 5 × 103 vgs/cell of the different AAV5 vectors. Forty-eight hours post-transduction, cell suspensions were analyzed for EGFP expression by flow cytometry. Representative histograms are shown. The data generated is from mean of triplicate analyses from two independent experiments. (b) Visual comparison of AAV5 S/T/K mutants in comparison to WT-AAV5 vectors. EGFP expression was detected by fluorescence microscopy 4-weeks post-administration of 5 × 1010 vector particles/animal of WT-AAV5 or mutant vectors. Representative images of hepatic tissues from four different animals in each group are shown. (c) Analysis of EGFP transcript levels by real-time quantitative PCR. Hepatic RNA isolated from animals injected with WT-AAV5 or AAV5 mutant vectors were analyzed for EGFP expression and the data normalized to the GAPDH reference gene. (d) Estimation of vector genome copies in the liver after AAV5 mediated gene transfer. Genomic DNA was isolated from liver tissue of C57BL/6 mice 4-weeks post vector administration and the viral copy numbers estimated by a quantitative PCR as described in the “materials and methods”. *p < 0.05 Vs WT-AAV5 injected animals.
In anticipation of achieving a robust gene expression from the AAV5 mutant vectors, we next examined their potential efficacy in C57BL/6 mice in vivo. All the seven AAV5 S/T/K mutant vectors expressing EGFP were administered at a dose of 5 × 1010 vgs/animal. Four-weeks after vector administration, mice administered with the three S → A (S268A, S652A and S658A) and one of the threonine mutant (T107A) showed higher levels of EGFP reporter when compared to animals injected with WT-AAV5 vector alone (Figure 1b). We then measured the transcript levels of EGFP in the hepatic RNA isolated from these mice. Our data demonstrates higher levels of EGFP transcripts (~4–16 fold) in AAV5-S/T mutant administered mice (Figure 1c). The highest transgene expression was noted from animals injected with AAV5 S652A vector, similar to the data from in vitro experiments. To validate these findings, we then measured the AAV vector genome copy numbers in various tissues of vector- or mock- injected mice. As shown in Figure 1d, a significant increase in the vector copies per diploid genome was observed in animals injected with S652A mutant vector (0.5 vs. 0.03, ~16 fold increase) in comparison to WT-AAV5 administered mice. Interestingly, some of the mutants (K32R, T328A, S485A) had lower infectivity in hepatocytes (Figure 1b and c). However, these mutant vectors were considerably re-targeted to other tissues such as the lungs (Supplementary Table S1), which may explain the low levels of EGFP expression seen in the liver. However, further studies are needed to confirm the mechanistic basis of re-targeting of some of these AAV5 mutant vectors, a phenomenon we have described earlier with AAV8 T252A vectors as well18.
Immunogenicity of AAV5 S/T mutant vectors is comparable to WT-AAV5 vectors
We then assayed neutralizing antibody formation in serially diluted serum samples from animals injected with AAV5 S268A, S485A, S652A, S658A and T107A vectors. Our data (Supplementary Table S2) does not show any change in NAb titers for the various groups of animals tested. Furthermore, the in vivo administration of these AAV5 S/T mutant vectors did not lead to any significant histological abnormalities in the livers of C57BL/6 mice 4 weeks post vector administration. Livers of mice injected with either WT-AAV5 or mutant vectors were grossly normal with comparable inflammation scores (Supplementary Table S3). Taken together, our results indicate that select S/T → A mutations can augment the transduction efficiency of AAV5 vectors in vivo and that these mutations also do not modify the immunogenic properties of the AAV5 capsid.
Serine and lysine mutant AAV1 vectors demonstrate increased transduction both in vitro and in vivo
The WT and each of the S/T/K mutant AAV1 capsids were packaged with a firefly luciferase transgene under the control of a chicken beta-actin promoter. Subsequently, they were evaluated for their in vitro transduction efficiency in CHO cells at a MOI of 5000 vgs/cell. The transgene expression level was measured 48 hours post-infection using luciferase reporter assay (BioVision, Milpitas, CA, USA). Of the seven AAV1 mutants tested, the luciferase activity measured as relative luminescence units (RLU) was significantly higher (~3.4–3.8 fold) from three of the vectors (AAV1- K137R, S663A and S669A) when compared to cells infected with the WT-AAV1 vector alone (Figure 2a). No further increase in the transduction efficiency was observed with the AAV1- S277A, S499A, T251A vectors as well as the control mutant S526A vector.
AAV1 serine/lysine mutant vectors demonstrate increased transduction efficiency in vitro and in vivo.
(a) In vitro transduction efficiency of AAV1 vectors. Equal number of CHO cells was mock-infected or infected with 5 × 103 vgs/cell of the different AAV1 vectors. Forty eight hours later, the luciferase activity in the cell lysate was measured as outlined in ‘Materials and Methods’. The data depicted are mean of triplicate analysis. Fold increase in transduction in comparison to WT-AAV1 infected cells are shown. *p < 0.05 Vs WT-AAV1 infected cells. (b) In vivo bio-luminescence imaging of AAV1 vector administered mice. Animals injected with 5 × 1010 vgs of AAV1-Luciferase vectors were imaged 2-weeks after gene transfer in an IVIS Spect-CT small animal imaging system (Perkin Elmer, Caliper Life Sciences). (c) The fold change in luciferase expression from animals injected with AAV1-K137R or AAV1-S669A vectors are shown in comparison to WT-AAV1 injected mice. *p < 0.05 Vs WT-AAV1 injected mice. (d) Ubiquitination profile of AAV1 vectors. Approximately 3 × 108 viral particles of WT-AAV1 and K137R-AAV1 vectors were denatured at 95°C for 5 minutes. The denatured viral particles were then used to perform the ubiquitin conjugation assay according to the manufacturer's protocol. The processed samples were electrophoresed on a 4–20% denaturing polyacrylamide gel and the ubiquitination pattern detected by immunoblotting. The mono-to-polyubiquitin conjugates are detected as a smear at molecular weight >150 Kda. (e) VP1 (87 Kda), VP2 (72 KDa) and VP3 (62 Kda) capsid proteins were immunoblotted as loading controls.
In view of the significant enhancement in the in vitro transduction efficiency of AAV1-mutants, it was of interest to examine their transduction potential in vivo. Approximately 5 × 1010 vgs of WT-AAV1 vector alone or the modified AAV1 (S669A, K137R) vector were injected into C57BL/6 mice intravenously. Phosphate-buffered saline (PBS)-injected mice were used as an appropriate control. Two-weeks later, an in vivo bioluminescence imaging was performed. Consistent with previously published studies10, animals injected with WT-AAV1 vectors demonstrated luciferase activity in the hepatic region. In comparison, while groups of animals administered with the AAV1-S669A vector had modestly elevated levels of luciferase activity (3 fold), animals injected with AAV1-K137R vector demonstrated substantially higher levels of gene expression (~6 fold) (Figure 2b, 2c). Further follow-up of these mice are needed to understand if this elevated gene expression is sustained over a period of time.
Finally, to investigate the basis for the improved transduction seen with AAV1- K137R vectors, we performed in vitro ubiquitination assays. As shown in Figure 2d, the AAV1- K137R vector had modestly reduced ubiquitination when compared to WT-AAV1 vector. These data corroborate that the superior transduction achieved with the lysine mutant AAV vectors across different serotypes, AAV2-K532R16, AAV8-K137R18 and the AAV1-K137R in the present study, is due to decreased capsid poly-ubiquitination which possibly results in rapid intra-cellular trafficking of the virus and consequently better gene expression.
Discussion
Despite significant advancements in therapeutic gene transfer using AAV vectors, it is well recognized that host and vector-related immune challenges need to be overcome for long-term gene transfer5. These factors have propelled the need to study the alternative AAV (1–10) serotypes further31,32. Each of these AAV1-10 serotype vectors possess unique biological characteristics conferred by a variable capsid sequence/structure23,33,34,35. Indeed, the use of AAV8 vectors successfully in hemophilia B clinical trials has demonstrated the utility of these alternate serotypes. However, it is also clear that one possibly needs to generate an AAV1-10 vector tool kit in view of pre-existing immunity against most of these serotypes and to target a larger proportion of patients. In addition, any further modifications to the native or wild-type AAV1-10 vectors that can improve their transduction efficiency will be desirable to counter vector dose-dependent immune response.
To address some of these issues, we have evaluated in the present studies the transduction efficiency of two bio-engineered alternate AAV serotype vectors 1 and 5. We hypothesized that selective modification of S/T/K residues within AAV1 or AAV5 capsid can improve their gene transfer as we have demonstrated successfully with AAV2 and AAV8 vectors16,18. In addition, AAV1 and AAV5 were specifically chosen for these studies as they are known to target liver efficiently and AAV5, in particular, has a low seroprevalence19,21. To the best of our knowledge, this is the first study that has selectively targeted phosphorylation sites in both AAV1 and AAV5 vectors to improve their phenotype. Limited studies have attempted to modify the capsid protein in AAV1 and AAV5 vectors. Wu et al, have demonstrated that swapping 6 dissimilar residues (L129, E418, E531, F584, A598 and N642) in the capsid of the closely-related AAV 1 and 6 affects the viral packaging titer, receptor binding and tissue tropism36. Similarly in case of AAV5 vectors, Hida et al, generated a random library of small deletions and tandem duplications, in which one of the duplications (dup 574_575) improved the transduction efficiency two-fold in BEAS-2B cells but not in 293-T cells in vitro37. The data from the present study indicates that selective mutation of potential Ser/Thr kinase or ubiquitination targets (S/T and K residues, respectively), in both AAV1 and AAV5 are beneficial. Some of these novel mutant AAV1 (K137R) and AAV5 (S652A) vectors had significantly higher (~2–16 fold vs. WT vectors) liver-directed gene expression in vivo. In addition, none of these novel AAV5 vectors showed any significant increase in hepatic inflammation as well as NAb titers in C57BL/6 mice when compared to the WT-AAV5 vector, which augurs well for their therapeutic use. On-going studies with the optimal mutant AAV1 (K137R) and AAV5 (S652A) vectors expressing human coagulation factor IX in preclinical models of hemophilia B, will demonstrate the feasibility of the use of these novel vectors for hepatic gene therapy. Interestingly, certain AAV5 mutant vectors like K32R, T328A and S485A were also considerably retargeted to other tissues, especially into the lungs (Supplementary Table S1). Such vectors may be useful in treating lung diseases such as cystic fibrosis airway disease and chronic obstructive pulmonary disease26,38,39.
S/T/K residues are abundant on AAV1 (18.8%) and AAV5 (18.5%) capsid. In the present studies we have shown proof- of- concept the efficacy of select, single- mutant AAV1 or AAV5 vectors. The availability of multiple combinations of these mutant vectors possibly encompassing other S/T/K targets within these capsids, is likely to further improve their efficiency and potentiate their evaluation in various disease states.
Methods
Cell lines and animals
Chinese hamster ovary (CHO) and AAV293 cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and Stratagene (Santa Clara, CA, USA) respectively. C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbour, ME, USA). All animal experiments were approved and carried out according to the Institutional guidelines for animal care (Christian Medical College, Vellore, India).
Site-directed mutagenesis
S/T → A, K → R mutations were introduced on the AAV1 and AAV5 rep/cap plasmid by QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's protocol. The primer sequences used are described in Supplementary table S4 and S5. The presence of the desired point mutation was verified by DNA sequencing (Life Technologies, Applied Biosystems 3130 Genetic Analyzer, Warrington, UK). The amino acid nomenclature on AAV capsid is according to NCBI database (accession number: AAV1- NC_002077.1, AAV5- NC_006152.1).
Generation of recombinant vectors
Highly purified stocks of self-complementary WT or the mutant AAV1 and AAV5 vectors containing either the EGFP or the firefly luciferase gene driven by the chicken β-actin promoter were generated by polyethyleneimine based triple- transfection of AAV-293 cells as described previously16. The physical particle titers of the vectors were quantified by slot blot analysis and expressed as viral genomes (vgs)/ml40.
Recombinant AAV vector transduction assays in vitro
To assess the efficacy of the novel mutant vectors generated, Chinese hamster ovary (CHO) cells were mock infected or infected with 5 × 103 vgs/cell of either WT- or the S/T/K mutant AAV1 and AAV5 vectors. Forty-eight hours post-transduction with AAV5-EGFP vectors, the transgene expression was measured by flow cytometry (FACS Calibur, BD, USA). In cells infected with AAV1-Luciferase vectors, the luciferase activity was measured using a commercial kit (BioVision Inc, Milpitas, CA, USA) in a GlowMax™ 20/20 luminometer (Promega, WI, USA). Each experiment was performed in triplicates and a mean of three independent experiments was used to assess change in transgene expression from different vectors evaluated.
Recombinant AAV vector transduction studies in vivo
Groups of 8–12 weeks-old C57BL/6 mice (n = 3–4 per group) were mock-injected or injected with 5 × 1010 vgs each of WT-AAV5 or WT-AAV1 and the corresponding S/T/K mutant vectors via the tail vein. Mice injected with AAV5-EGFP vectors were euthanized 4-weeks after vector administration. Cross sections from hepatic lobes of all animals were assessed for EGFP expression in a fluorescence microscope (Leica CTR6000, GmbH, Germany). Animals injected with AAV1-luciferase vectors were imaged for bio-luminescence in an IVIS Spect-CT small animal imaging system (Perkin Elmer, Caliper Life Sciences, Hopkinton, MA). D-Luciferin (BioVision Inc; 2.5 mg in 100 μL for 20 gm mouse body weight) was injected intraperitoneally. Images were acquired at 10 min after injection.
Real-time PCR assays
To measure EGFP transcript levels in AAV5 administered mice, total RNA was isolated from the murine liver samples 4 weeks post-vector administration (5 × 1010 vgs per mouse) using TRIZOL® reagent (Sigma Aldrich, St. Louis, MO, USA). Approximately 1 μg of RNA was reverse-transcribed using Verso™ Reverse Transcriptase (Thermo Scientific, Surrey, UK). TAQMAN® PCR was done using primers/probe against EGFP gene16 according to the manufacturer's protocol (Eurogentec, Seraing, Belgium). GAPDH was used as the housekeeping control gene. Data was captured and analyzed using the ABI Prism 7500 Sequence Detection (Life Technologies, Applied Biosystems).
To study the bio-distribution of AAV5 vectors in recipient mice, various tissues such as liver, spleen, lung, heart, kidney and muscle tissue were collected, 4 weeks after gene transfer. Genomic DNA was used to estimate the vector copy numbers in 100 ng of template genomic DNA by amplifying the viral inverted terminal repeats (ITRs) with specific probes/primers as described previously41 in a PCR mastermix according to manufacturer's protocol (Eurogentec). Data was captured and normalized to mouse GAPDH housekeeping control gene and analyzed in the ABI Prism 7500 Sequence Detection System (Life Technologies).
Ubiquitin conjugation assay and immunoblotting
Ubiquitination assay was performed using the Ubiquitin-Protein Conjugation kit (Boston Biochem, MA, USA). The ubiquitination pattern was detected by immunoblotting of the samples with mouse anti-ubiquitin monoclonal antibody (P4D1) and HRP-conjugated anti-mouse IgG1 secondary antibody (Cell Signaling Technology, Boston, MA, USA). To check for equal loading of the samples, VP1, VP2 and VP3 capsid proteins were detected by AAV clone B1 antibody (Fitzgerald, North Acton, MA, USA) and HRP-conjugated anti-mouse IgG1 secondary antibody (Cell Signaling Technology).
Statistical analysis
Data are expressed as mean ± standard deviation, unless otherwise indicated. Multiple comparisons between groups were performed with analysis of variance (ANOVA) tests. *p < 0.05 denotes statistical significance.
References
Bennett, J., Ashtari, M., Wellman, J., Marshall, K. A., Cyckowski, L. L., Chung, D. C. et al. AAV2 gene therapy readministration in three adults with congenital blindness. Sci Transl Med 4, 120ra115 (2012).
Bartel, M. A., Weinstein, J. R. & Schaffer, D. V. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther 19, 694–700 (2012).
Grieger, J. C. & Samulski, R. J. Adeno-associated virus as a gene therapy vector: vector development, production and clinical applications. Adv Biochem Eng Biotechnol 99, 119–145 (2005).
Daya, S. & Berns, K. I. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 21, 583–593 (2008).
High, K. A. The gene therapy journey for hemophilia: are we there yet? Blood 120, 4482–4487 (2012).
Manno, C. S., Pierce, G. F., Arruda, V. R., Glader, B., Ragni, M., Rasko, J. J. et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med 12, 342–347 (2006).
Buchlis, G., Podsakoff, G. M., Radu, A., Hawk, S. M., Flake, A. W., Mingozzi, F. et al. Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood 119, 3038–3041 (2012).
Mingozzi, F., Maus, M. V., Hui, D. J., Sabatino, D. E., Murphy, S. L., Rasko, J. E. et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med 13, 419–422 (2007).
Madsen, D., Cantwell, E. R., O'Brien, T., Johnson, P. A. & Mahon, B. P. Adeno-associated virus serotype 2 induces cell-mediated immune responses directed against multiple epitopes of the capsid protein VP1. J Gen Virol 90, 2622–2633 (2009).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16, 1073–1080 (2008).
Hu, C. & Lipshutz, G. S. AAV-based neonatal gene therapy for hemophilia A: long-term correction and avoidance of immune responses in mice. Gene Ther 19, 1166–1176 (2012).
Maheshri, N., Koerber, J. T., Kaspar, B. K. & Schaffer, D. V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol 24, 198–204 (2006).
Finn, J. D., Hui, D., Downey, H. D., Dunn, D., Pien, G. C., Mingozzi, F. et al. Proteasome inhibitors decrease AAV2 capsid derived peptide epitope presentation on MHC class I following transduction. Mol Ther 18, 135–142 (2010).
Mingozzi, F., Hasbrouck, N. C., Basner-Tschakarjan, E., Edmonson, S. A., Hui, D. J., Sabatino, D. E. et al. Modulation of tolerance to the transgene product in a nonhuman primate model of AAV-mediated gene transfer to liver. Blood 110, 2334–2341 (2007).
Nathwani, A. C., Tuddenham, E. G., Rangarajan, S., Rosales, C., McIntosh, J., Linch, D. C. et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365, 2357–2365 (2011).
Gabriel, N., Hareendran, S., Sen, D., Gadkari, R. A., Sudha, G., Selot, R. et al. Bio-engineering of AAV-2 capsid at specific serine, threonine or lysine residues improves its transduction efficiency in vitro and in vivo. Hum Gene Ther Methods (2013). EPUB doi: 10.1089/hgtb.2012.194.
Zhong, L., Li, B., Mah, C. S., Govindasamy, L., Agbandje-McKenna, M., Cooper, M. et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci U S A 105, 7827–7832 (2008).
Sen, D., Gadkari, R. A., Sudha, G., Gabriel, N., Sathish Kumar, Y., Selot, R. et al. Targeted modifications in adeno-associated virus (AAV) serotype -8 capsid improves its hepatic gene transfer efficiency in vivo. Hum Gene Ther Methods (2013). EPUB doi: 10.1089/hgtb.2012.195.
Boutin, S., Monteilhet, V., Veron, P., Leborgne, C., Benveniste, O., Montus, M. F. et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8 and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21, 704–712 (2010).
Calcedo, R., Vandenberghe, L. H., Gao, G., Lin, J. & Wilson, J. M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199, 381–390 (2009).
Li, C., Narkbunnam, N., Samulski, R. J., Asokan, A., Hu, G., Jacobson, L. J. et al. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther 19, 288–294 (2012).
Murphy, S. L., Li, H., Zhou, S., Schlachterman, A. & High, K. A. Prolonged susceptibility to antibody-mediated neutralization for adeno-associated vectors targeted to the liver. Mol Ther 16, 138–145 (2008).
Paneda, A., Vanrell, L., Mauleon, I., Crettaz, J. S., Berraondo, P., Timmermans, E. J. et al. Effect of adeno-associated virus serotype and genomic structure on liver transduction and biodistribution in mice of both genders. Hum Gene Ther 20, 908–917 (2009).
Xiao, W., Chirmule, N., Berta, S. C., McCullough, B., Gao, G. & Wilson, J. M. Gene therapy vectors based on adeno-associated virus type 1. J Virol 73, 3994–4003 (1999).
Lorain, S., Gross, D. A., Goyenvalle, A., Danos, O., Davoust, J. & Garcia, L. Transient immunomodulation allows repeated injections of AAV1 and correction of muscular dystrophy in multiple muscles. Mol Ther 16, 541–547 (2008).
Zabner, J., Seiler, M., Walters, R., Kotin, R. M., Fulgeras, W., Davidson, B. L. et al. Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J Virol 74, 3852–3858 (2000).
Hildinger, M., Auricchio, A., Gao, G., Wang, L., Chirmule, N. & Wilson, J. M. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J Virol 75, 6199–6203 (2001).
Nathwani, A. C., Gray, J. T., McIntosh, J., Ng, C. Y., Zhou, J., Spence, Y. et al. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood 109, 1414–1421 (2007).
Halbert, C. L., Miller, A. D., McNamara, S., Emerson, J., Gibson, R. L., Ramsey, B. et al. Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5 and 6 in cystic fibrosis and normal populations: Implications for gene therapy using AAV vectors. Hum Gene Ther 17, 440–447 (2006).
Gaudet, D., Methot, J., Dery, S., Brisson, D., Essiembre, C., Tremblay, G. et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPL(S447X)) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther (2012).
Zincarelli, C., Soltys, S., Rengo, G., Koch, W. J. & Rabinowitz, J. E. Comparative cardiac gene delivery of adeno-associated virus serotypes 1–9 reveals that AAV6 mediates the most efficient transduction in mouse heart. Clin Transl Sci 3, 81–89 (2010).
Gao, G., Vandenberghe, L. H. & Wilson, J. M. New recombinant serotypes of AAV vectors. Curr Gene Ther 5, 285–297 (2005).
Wu, Z., Asokan, A. & Samulski, R. J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther 14, 316–327 (2006).
Rebuffat, A., Harding, C. O., Ding, Z. & Thony, B. Comparison of adeno-associated virus pseudotype 1, 2 and 8 vectors administered by intramuscular injection in the treatment of murine phenylketonuria. Hum Gene Ther 21, 463–477 (2010).
Markakis, E. A., Vives, K. P., Bober, J., Leichtle, S., Leranth, C., Beecham, J. et al. Comparative transduction efficiency of AAV vector serotypes 1–6 in the substantia nigra and striatum of the primate brain. Mol Ther 18, 588–593 (2010).
Wu, Z., Asokan, A., Grieger, J. C., Govindasamy, L., Agbandje-McKenna, M. & Samulski, R. J. Single amino acid changes can influence titer, heparin binding and tissue tropism in different adeno-associated virus serotypes. J Virol 80, 11393–11397 (2006).
Hida, K., Won, S. Y., Di Pasquale, G., Hanes, J., Chiorini, J. A. & Ostermeier, M. Sites in the AAV5 capsid tolerant to deletions and tandem duplications. Arch Biochem Biophys 496, 1–8 (2010).
Aneja, M. K. & Rudolph, C. Gene therapy of surfactant protein B deficiency. Curr Opin Mol Ther 8, 432–438 (2006).
Flotte, T. R., Ng, P., Dylla, D. E., McCray, P. B., Jr, Wang, G., Kolls, J. K. et al. Viral vector-mediated and cell-based therapies for treatment of cystic fibrosis. Mol Ther 15, 229–241 (2007).
Kube, D. M. & Srivastava, A. Quantitative DNA slot blot analysis: inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res 25, 3375–3376 (1997).
Aurnhammer, C., Haase, M., Muether, N., Hausl, M., Rauschhuber, C., Huber, I. et al. Universal Real-Time PCR for the Detection and Quantification of Adeno-Associated Virus Serotype 2-Derived Inverted Terminal Repeat Sequences. Hum Gene Ther 23, 18–28 (2012).
Acknowledgements
We thank Dr. R. Sumathy and Mr. Sathish Y., Laboratory animal core facility, Centre for Stem Cell Research, Vellore for animal care. GRJ is supported by research grants from Department of Science of Technology, Government of India (Swarnajayanti Fellowship 2011), Department of Biotechnology (DBT), Government of India (Innovative Young Biotechnologist award 2010: BT/03/IYBA/2010; Grant: BT/PR14748/MED/12/491/2010; Grant: BT/01/COE/08/03) and an early career investigator award-2010 from Bayer Hemophilia Awards program, Bayer Inc, USA. We thank Immunology Core, Gene Therapy Program, Department of Pathology and Laboratory Medicine, University of Pennsylvania, USA for estimation of neutralizing antibodies against AAV5.
Author information
Authors and Affiliations
Contributions
G.R.J. designed the experiments. D.S., B.B., N.G., P.A., R.S. and V.R. performed the experiments. D.S., B.B. and G.R.J. analyzed and interpreted the data. A.S. provided reagents and tools. D.S., B.B. and G.R.J. wrote the paper. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary file
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sen, D., Balakrishnan, B., Gabriel, N. et al. Improved adeno-associated virus (AAV) serotype 1 and 5 vectors for gene therapy. Sci Rep 3, 1832 (2013). https://doi.org/10.1038/srep01832
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01832
This article is cited by
-
Evaluating Gene Therapy as a Potential Paradigm Shift in Treating Severe Hemophilia
BioDrugs (2023)
-
Choice of vector and surgical approach enables efficient cochlear gene transfer in nonhuman primate
Nature Communications (2022)
-
Tissue and cell-type-specific transduction using rAAV vectors in lung diseases
Journal of Molecular Medicine (2021)
-
Future of Haemophilia Research in India
Indian Journal of Hematology and Blood Transfusion (2020)
-
Astrocyte-selective AAV gene therapy through the endogenous GFAP promoter results in robust transduction in the rat spinal cord following injury
Gene Therapy (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.