Abstract
Bacterial cell wall is targeted by many antibiotics. Among them are lantibiotics, which realize their function via interaction with plasma membrane lipid-II molecule — a chemically conserved part of the cell wall synthesis pathway. To investigate structural and dynamic properties of this molecule, we have performed a series of nearly microsecond-long molecular dynamics simulations of lipid-II and some of its analogs in zwitterionic single component and charged mixed simulated phospholipid bilayers (the reference and the mimic of the bacterial plasma membrane, respectively). Extensive analysis revealed that lipid-II forms a unique “amphiphilic pattern” exclusively on the surface of the simulated bacterial membrane (and not in the reference one). We hypothesize that many lantibiotics exploit the conserved features of lipid-II along with characteristic modulation of the bacterial membrane as the “landing site”. This putative recognition mechanism opens new opportunities for studies on lantibiotics action and design of novel armament against resistant bacterial strains.
Similar content being viewed by others
Introduction
Many organisms possess cell wall on the outside from the plasma membrane — archaea, bacteria, fungi and plants. Peptidoglycan-built cell wall is crucial for survival of bacteria and that is why it is considered to be the target for next generation of antibiotics capable of overcoming the effect of multi-resistance1,2. Various classes of bactericidal substances inhibit cell wall growth. Among them is the most known and first widely-used antibiotic penicillin which blocks transpeptidase what results in disruption of the peptidoglycan layer. Another class is lantibiotics — ribosomally synthesized peptides, which are produced by gram-positive bacteria and undergo several common post-translational modifications such as introduction of intramolecular lanthionine and methyllanthionine cycles3,4.
Apart from penicillin, lantibiotics disturb transport of the cell wall elementary units from the cytoplasm to the membrane surface via interactions with lipid-II5,6 (Fig. 1A) — a “shuttle carrier” of peptidoglycan “building blocks” which consists of this block and a long bacterioprenol hydrophobic chain connected with pyrophosphate linker. It has been demonstrated that lantibiotics cause dual effect: selectively trap lipid-II and inhibit cell wall growth and/or disrupt the plasma membrane exploiting lipid-II as a high-affinity anchor7.
MD simulations of lipid-II in PG/PE membrane.
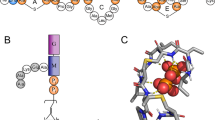
(A) Chemical structure of lipid-II: peptidoglycan “building block” (shown with colored symbols) is connected via pyrophosphate moiety (orange circles) to bacterioprenol hydrophobic chain, which consists of eleven isoprenoid monomers. Lipid-III in contrast to lipid-II does not have pentapeptide moiety. “Short” lipid-II analog used in our MD-simulations has a tail comprised of only four isoprene segments. (B) Typical conformation of lipid-II in PG/PE membrane during MD simulation: tail (yellow) is “L-shaped”, pyrophosphate (orange) is partially buried into membrane–water interface and disaccharide (cyan) is “embraced” with pentapeptide (pink). (C) “V-shaped tail” of lipid-II (the terminal group is able to emerge to the water–lipid interface). (D) Density profiles of various lipid-II groups (left axis) and solvent (violet; right axis). Results are shown for the following lipid-II groups: peptide (pink), sugar residues (cyan), pyrophosphate (orange), tail (yellow) and the terminal segment of the tail (S-11; brown). (E) Hydrogen-bonding pattern within lipid-II head group and with phospholipids' polar groups. Blue broken lines depict hydrogen bonds between head group monomers (larger thickness corresponds to high occupancy of hydrogen bonds in MD). Symbols' color indicates whether monomer is hydrogen bond donor (blue), acceptor (green), or both (magenta). Bold symbol outline points monomers that form extensive hydrogen bonding with the membrane (and therefore have low relative solvent accessibility, see Fig. S1).
Lipid-II is a low-abundant molecule (less than 1 mol % of membrane phospholipids (PLs)8), which performs the cycle of peptidoglycan “building block” translocation in less than 1 second. Lipid-II is found mostly in regions of plasma membrane related to cell division and elongation6, although it does not tend to aggregate itself, display homogeneous distribution in giant unilamellar vesicles (GUV)7 and spontaneously partition into fluid domains of two-component lipid bilayers9. Atomic-force microscopy (AFM) studies of such lipid-II–containing bilayers show that the head group of lipid-II is approximately 1.9 nm high as compared to the membrane surface; although, it is rather soft and can be easily and reversibly penetrated by the AFM probe9.
The principal structural motif of lipid-II recognized by lantibiotics is shown to be pyrophosphate and recognition itself occurs at the extracellular surface of the membrane10. Structure of nisin–lipid-II complex obtained by nuclear magnetic resonance (NMR) spectroscopy in DMSO solution contains unique “pyrophosphate cage”11,12 comprised of nisin lanthionine rings, where altering L- and D-amino acids form a nest-like motif13. It's important to note that DMSO is very rough mimic of the membrane, so the actual structure of this complex in the membrane may be different. Several other lantibiotics and antimicrobial peptides, including mersacidin14, ramaplanin15, katanosin16, plectasin17 and others, bind to pyrophosphate as well, although interaction sites most probably differ from each other and typically embrace one or both sugar residues of peptidoglycan part. Although much effort has been spent for rationalization of structure–activity relationships of lantibiotics18, molecular aspects of their action are still unclear, especially when it comes to real bacterial membranes. For instance, it is difficult to understand how lantibiotics find lipid-II molecules in the “sea” of phospholipids and selectively capture them. At the same time, it was shown that lipid-II is a nisin-specific target in bacterial membranes and that addition of lipid-II to simulated membrane systems increases affinity of nisin by three orders of magnitude (to nanomolar range)19. On this basis, the problem of lipid-II recognition by extracellular compounds is very challenging, since lipid-II has rather constant chemical structure and therefore represents suitable target for design of novel antibiotics1,5.
Being complementary to experimental studies, molecular modeling gives an opportunity to address the problem of lipid-II recognition by lantibiotics with an atomic resolution, although little effort has been made to achieve this. The sole accomplished work describes binding of vancomycin to lipid-II in PL bilayer20, but little is said about founding principles of this recognition and the dynamic and structure of the target — lipid-II molecule. The continuation of this work pays more attention to dimerization of vancomycin and its binding to peptidoglycan analogues and does not cover issue of membrane environment of lipid-II21.
The main objective of the present study is to understand at the molecular level, what makes lipid-II “visible” to lantibiotics in the crowded phospholipid environment of bacterial membranes. With this aim in view we have performed a series of nearly microsecond-long molecular dynamics (MD) calculations of lipid-II and its several analogs in the simulated bacterial membrane mimic and in the reference zwitterionic bilayer. The following questions regarding to the structure of lipid-II and the surrounding membrane were assessed: 1) What conformations and dynamic behavior are peculiar to lipid-II embedded in lipid bilayers of different composition — resembling bacterial and eukaryotic membranes? 2) Whether or not lipid-II creates any particular “pattern” in the neighboring membrane areas, which can be recognized by extracellular lantibiotics? 3) Do two lipid-II molecules dimerize in the membrane and whether this may be a prerequisite for lantibiotics' recognition of bacterial membrane? In conclusion, we discuss possible mechanism of capture of lipid-II by lantibiotic peptides.
Results
In this study we explore conformational properties of lipid-II in bacterial membranes with the help of full-atom MD simulations. To mimic membranes of gram-positive bacteria, we use a 3:1 mixture of palmitoyloleoylphosphatidylglycerol (POPG) with palmitoyloleoylphosphatidylethanolamine (POPE) (hereinafter, this membrane will be referred to as PG/PE)22. These lipids are the major anionic and zwitterionic species in bacteria, respectively. While 1:3 PG/PE is a good mimic for gram-negative membrane (such as E. coli23), 3:1 is more like gram-positive one24. As a reference zwitterionic membrane we use pure palmitoyloleoylphosphatidylcholine (POPC) bilayer. A number of MD trajectories, each of 0.4–1 μs, were calculated for the following systems: 1) pure POPC; 2) pure PG/PE; 3) POPC/lipid-II; 4) PG/PE/lipid-II; 5) PG/PE/lipid-III; 6) PG/PE/“short-tail lipid-II” and 7) PG/PE/two lipid-II molecules (see Methods and Table 1). “Truncated” analogs of lipid-II — lipid-III and “short-tail” lipid-II (see Fig. 1A for structures) — were used to assess particular roles of the lipid-II head and the bacterioprenol tail groups in its dynamic behavior and effect on the simulated lipid bilayers.
The main stages of the research were the following: 1) MD calculations and basic analysis for the mentioned systems; 2) Assessment of lipid-II (or its analogs) conformational dynamics and inter-/intramolecular interactions; 3) Description of the “membrane response” (2D-maps of hydrophobic properties and lipid chains' order parameters, analysis of clusters that are induced in the membrane); 4) Characterization of interaction of two lipid-II molecules in PG/PE bilayer.
Lipid-II in bacterial membrane is highly flexible
In the MD trajectory of lipid-II in PG/PE membrane we observed that pyrophosphate (PP) group behaves like an “anchor”, which binds strongly to the lipid–water interface (see below). The lipid-II head group “floats” above the bilayer surface, while the tail maintains itself inside the hydrophobic core of the simulated membranes.
The head group: a dynamic “anchor”
The head group of lipid-II is peptidoglycan “building block” of the cell wall, which consists of two sugar residues (N-acetylmuramic acid (M) and N-acetylglucosamine (G)) and modified 5-mer peptide connected via linker to M sugar residue (for lipid-II structure see Fig. 1A). In the course of MD, disaccharide moiety resides right above PP group with G residue “capping” M residue that results in maximum relative solvent accessibility (>80%) of G as compared to M and any other part of the head group (see Supplementary Fig. S1 online). The first three residues of the peptide moiety “wrap” the disaccharide and form an extensive hydrogen-bonding network with G-sugar (Fig. 1E). In addition, two peptide residues — γ-D-glutamyl and lysyl — also display strong H-bonding with phospholipids being both donors and acceptors. Furthermore, strong electrostatic attraction of Lys positive charge to negatively charged PG/PE surface makes this residue relatively inaccessible (only ≈25%) to the solvent (Supplementary Fig. S1 online). The C-terminal dipeptide — D-alanyl–D-alanine — is the most solvent-accessible (70–80%) and flexible part of the head group (the root-mean-square fluctuation monotonically increases from 0.3 to 0.6 nm per residue within the pentapeptide). From the dynamic point of view, this dipeptide “shields” first three residues and partially the disaccharide like an umbrella (Fig. 2). The maximum elevation of the C-terminus above the membrane surface is ≈2 nm, although the mean “height” of the head group is much lower (see below). The “shielding” function of the pentapeptide leaves accessible to the solvent N-acetyl and –CH2–OH groups of both sugar residues.
Dynamic states density of lipid-II in PG/PE membrane.
Lipid-II conformations are extracted from the equilibrium part (30–1000 ns) of MD trajectory with time step of 2 ns, as viewed from the membrane (A) and extracellular side (B). Gray spheres correspond to PLs' phosphorus atoms from a single MD frame and denote the water–membrane interface; PL atoms from other frames, water and ions are not shown for clarity. Lipid-II groups are colored as follows: tail, sandy; pyrophosphate, black; sugar M, red; sugar G, orange; Ala, green; γ-D-Glu, cyan; Lys, blue; 1st D-Ala, magenta; 2nd D-Ala, purple. Pink spheres show C-terminus of the pentapeptide. A, lower panel: Radial distribution function (RDF) of lipid-II tail terminus (S-11) with respect to PP. RDF is truncated at distance 3 nm. Note that this graph is symmetrical.
Density profiles of lipid-II atom groups in PG/PE bilayer (Fig. 1D) show that: 1) the peptide and the sugar moieties float above the membrane–water interface, with sugar G and D-Ala–D-Ala C-terminal dipeptide being the most solvent-accessible (Supplementary Fig. S1 online); 2) PP group resides slightly deeper (Fig. 1B) and “anchors” the molecule to the membrane–water interface; 3) the tail has relatively sharp distribution with a maximum in the center of the membrane, while 4) the terminal tail segment (11th monomer S-11) is able to localize in any layer of the membrane (Fig. 2A).
The tail group: a “mixing bar” in lipid membrane milieu
The first half of the long bacterioprenol tail of lipid-II (starting from PP) penetrates inside towards the middle of the membrane and adopts an average angle from the bilayer normal of 128 ± 15° (Supplementary Fig. S2 online). The length of the 11-mer bacterioprenol chain is sufficient to span even the whole membrane that determines dynamics of the second half of the tail. The most typical MD conformation of lipid-II tail is “L-shaped” (Fig. 1B), where its second half resides between two monolayers. At the same time, the tail is extremely flexible and it behaves like a “mixing bar”, spanning 5 nm vicinity of the head group (Fig. 2). The terminal tail segment also quite often emerges at the opposite (fully extended tail conformation, “I-shaped”) or even at the same surface of the membrane. In the latter case, the “V-shaped” (Fig. 1C) tail is able to push the phospholipids tails aside and may reach the surface of the bilayer (despite being deeply buried inside the hydrophobic core of the membrane, the tail still has non-zero value of MD-averaged solvent-accessible area, see Supplementary Fig. S1 online). The relative MD occupancies of “L-shaped”, “V-shaped” and “I-shaped” conformations of lipid-II tail in PG/PE bilayer are 0.70, 0.12 and 0.18, respectively (see Supplementary Fig. S3 online for details).
Highly flexible lipid-II tail disturbs phospholipid acyl chains that is evident from calculations of their deuterium order parameters (Scd, see Methods). Figure 3B shows local disordering of PG/PE bilayer in the area where the tail is preferentially located. Pure PG/PE membrane shows more homogeneous in-plane Scd distribution (Fig. 3A), while integral Scd values are very similar for the bilayers with and without lipid-II (data not shown).
Lipid-II induces amphiphilic pattern in PG/PE membrane.
(A) Phospholipid acyl chains order parameters (Scd) of pure PG/PE bilayer mapped on the membrane surface. Color represents MD averaged Scd values for both PL acyl chains: from green (disordered chain) to violet (relatively ordered chain). (B) Scd map for PG/PE bilayer with lipid-II (red). Note extended disordered area next to lipid-II molecule. (C) 2D-map of hydrophobic properties of pure PG/PE membrane (view from the “top” corresponding to panel A). Hydrophobicity is represented by the Molecular Hydrophobicity Potential (MHP), which was calculated at bilayer surface, mapped in the plane and MD-averaged. Colored circles depict projections of POPG (blue) and POPE (green) phosphorus atoms. (D) MHP-map of PG/PE bilayer with lipid-II (view corresponds to panel B). Lipid-II is shown with yellow triangles (tail segments), orange circles (phosphorus atoms), cyan circles (sugar residues) and pink circles (amino acid residues). Colored contours show areas of the most probable location of lipid-II atomic groups during MD (colored according to symbols). Note prominent hydrophobic “atoll” surrounding lipid-II which is co-localized with lipid-II tail group (marked with broken line) and partially corresponds to the disordered area in panel B. Results of the same analysis of POPC bilayer properties (pure and with lipid-II) can be found in Fig. S5.
Lipid-II creates amphiphilic pattern on the surface of bacterial membrane
Large-amplitude motions of lipid-II tail (Fig. 2) significantly disturb lipid phase of the PG/PE membrane (see previous paragraph). To inspect these perturbations, we employed an original method of mapping of hydrophobic properties that lipids create on the membrane surface (see Methods).
“Atoll-like” pattern
MD-averaged hydrophobicity maps reveal amphiphilic “atoll-like” pattern that wraps the lipid-II molecule (Fig. 3D). This pattern consists of horseshoe-shaped hydrophobic surface (approx. 7 nm2) and a hydrophilic area inside it (approx. 5.5 nm2). In the course of MD, this “atoll” is formed by several (dis)continuous patches of hydrophobic surface (see Supplementary video S1), acquiring the aforementioned shape as a result of time-averaging (during 900 ns time interval). The hydrophilic “knob” inside of this “atoll” is formed by lipid-II head group (Supplementary Fig. S4 online). According to radial profile of the bilayer landscape (with the center on the lipid-II molecule), lipid-II in PG/PE membrane appears as a knob with elevation of 0.45 ± 0.32 nm and cross-section radius at the half-height of 0.9 nm (data not shown). The maximum elevation of about 2 nm was observed during MD for lipid-II with extended pentapeptide conformation, where the C-terminus points to the solvent (Fig. 2). Even beyond lipid-II vicinity the surface of PG/PE bilayer remains highly disturbed: the landscape profile is very rough, up to 4 nm away from lipid-II.
Highly polar head group of lipid-II acts as both donor and acceptor of hydrogen bonds with phospholipid polar heads. In addition, positively charged Lys residue of the peptide moiety electrostatically attracts negatively charged lipids. By virtue of these interactions, lipid-II head group efficiently “traps” several PL molecules, which remain bound during MD simulations. This fact is illustrated by radial distribution functions (RDFs; see Fig. 4), where the first peak corresponds to tightly bound PL molecules (mostly POPE) and the following minimum indicates the “atoll”, which has decreased density of PL head groups. Evolution of lipid-II–POPE RDF over MD time is presented in Supplementary video S3, where the sharp peak is forming starting from rather more uniform distribution.
Radial distribution functions (RDF) for phosphate groups of various phospholipids: POPC (black) or POPG (blue) and POPE (green) with respect to lipid-II PP group.
These RDFs are calculated for the last 300 ns of MD trajectory. Inset: RDF for PPlipid-II–PPE in PG/PE membrane for the first 50 ns of MD. The dynamic RDF picture is shown in video S3. Dashed line corresponds to RDF = 1.
Hydrophobic surface of the “atoll” is formed by phospholipid acyl chains, which are partially expelled to the surface of the bilayer by the tail of lipid-II (due to local disordering). Our analysis testifies that the surface of PG/PE has more hydrophobic area in presence of lipid-II than in the pure bilayer (data not shown). The hydrophobic surface (“atoll”) emerges in the area of the most probable location of the lipid-II tail (see yellow outline in Fig. 3D). In the “atoll” area PLs' head groups are located sparsely, promoting exposure of acyl chains. At the same time, lipid-II head group tends to localize in the opposite side from the tail (pink outline in Fig. 3D), where the terrain is split by the “knob”. Note that maximal density of the tail states is observed in ≈120° region around PP with respect to peptide's maximal density of states (Fig. 2). By contrast, pure PG/PE membrane does not have such distinct and long-living hydrophobic patterns (Fig. 3C). Although being mosaic according to distribution of hydrophobic/hydrophilic properties (see Supplementary video S2), the surface of pure membrane is too flexible and dynamic to be able to maintain such long-living heterogeneities.
Hydrophobic clusters on the membrane surface
For more in-depth view of lipid-II–induced membrane heterogeneities we performed analysis of size (Fig. 5A) and lifetime (Fig. 5B) of hydrophobic clusters on the membrane surface (see Methods). It is evident that hydrophobic clusters are larger and live longer in PG/PE membrane with the presence of lipid-II (Fig. 5A, green curve), as compared to pure PG/PE bilayer. The most extended clusters (that have area >2.5–3 nm2) are obviously the same that form hydrophobic “atoll” around the lipid-II molecule. Almost identical distribution comes from the lipid-III–containing system (a lipid-II analogue with “truncated” peptide moiety in the head group) bilayer (Fig. 5A, red curve). Same conclusions may be drawn from presentation of cluster size distribution as a complementary cumulative distribution function (see Supplementary Fig. S6 online). Systems with lipid-II and lipid-III have very similar cluster lifetime distributions, as well (Fig. 5B).
Lipid-II induces formation of large and long-living hydrophobic clusters in PG/PE membrane and destroys them in POPC membrane.
(A) Distribution of cluster size over MD trajectories of six systems: pure POPC (black broken line), POPC+lipid-II (gray line), pure PG/PE (green broken line), PG/PE+lipid-II (green line), PG/PE+lipid-III (red line) and PG/PE+short lipid-II (yellow line). X-axis: cluster size (in nm2). Y-axis: Probability Density Function (PDF). Note that the largest clusters are observed in PG/PE with full-length tail molecule (lipid-II or -III). At the same time, the smallest clusters are observed in POPC+lipid-II system. Alternative analysis of cluster sizes (complementary cumulative PDF) is shown in Fig. S6. (B) Life-time distribution for hydrophobic clusters. X-axis is cluster lifetime (in ns). Y-axis is total lifetime for all clusters with the current lifetime or longer (in μs).
Lipid-II does not demonstrate any unique features in the reference zwitterionic membrane (POPC)
To understand whether or not lipid-II displays particular behavior in its “natural” lipid environment (e.g., in our anionic PG/PE bacterial membrane mimic), we have performed MD simulations of this molecule incorporated into zwitterionic POPC bilayer, which is often used in experiments and simulations. Similar analysis as described above gives the following important observations.
Lipid-II head group orientation is largely different in POPC as compared to PG/PE. Generally speaking, it submerges deeper under the membrane surface (compare density profiles in Supplementary Fig. S7 online and Fig. 1D) and is more oblique with respect to the membrane normal. Mean elevation of lipid-II head group above the POPC surface is 0.19 ± 0.26 nm (compare to ≈0.45 nm in PG/PE). The most pronounced difference is observed for G sugar residue and C-terminal D-Ala–D-Ala dipeptide, which are considerably buried into the lipid interior of POPC bilayer and have no more than 20% of solvent-accessible surface area as compared to >70% in PG/PE (Supplementary Fig. S1 online). At the same time, PP group of lipid-II in POPC bilayer partially resurfaces, as well as M sugar residue and the first three residues of pentapeptide moiety do. Consistently, head group obliquity angle (the angle between bilayer normal and head group major axis) is closer to right angle in POPC, while it is acute (~55°) in case of PG/PE (Supplementary Fig. S4 online).
This fact one can clearly get from bilayer surface landscape maps, where the lipid-II head group is more elevated in PG/PE with respect to the water–lipid interface than in POPC bilayer (compare panels A and B in Supplementary Fig. S3 online). Possible explanation of these differences can be lack of H-bond donors in POPC head groups, which decreases efficiency of interactions between phospholipid polar heads with each other and with lipid-II head as compared to PG/PE membrane (Supplementary Fig. S7 online).
Lipid-II tail also behaves differently in the hydrophobic core of POPC bilayer: it resides in more “vertical” orientation having the angle between bilayer normal and the first half of lipid-II tail of 151 ± 17°, in contrast to ≈128° in case of PG/PE (Supplementary Fig. S2 online). At the same time, POPC molds MD population of “L-shaped”, “V-shaped” and “I-shaped” conformations of lipid-II tail as 0.72, 0.17 and 0.11, respectively (compare to PG/PE's 0.70/0.12/0.18). The most pronounced difference is peculiar to the lower portion of “I-shaped” conformation that “pierces” the membrane (Supplementary Fig. S3 online).
Finally, apart from PG/PE membrane, no distinct amphiphilic pattern is observed in POPC bilayer (Supplementary Fig. S5 online). Being more flexible than PE/PG membrane, POPC still has rather “mosaic” hydrophilic/hydrophobic surface, but hydrophobic clusters do not have persistent positions in relation with lipid-II, nor is long lifetime observed for hydrophobic clusters in this bilayer with or without lipid-II. Moreover, lipid-II decreases acyl chains order parameters, as compared to pure POPC (Supplementary Fig. S5 online). Cluster analysis reveals that pure POPC has smaller and more short-living hydrophobic clusters, as compared to pure PG/PE (gray broken line in Fig. 5A), while incorporation of lipid-II into POPC membrane further breaks and destabilizes the clusters (see also Supplementary Fig. S6 online).
Role of lipid-II's head and tail groups in formation of the membrane surface pattern
To reveal particular roles of lipid-II head and tail groups in its intrinsic dynamics and modification of membrane properties, two lipid-II analogs that lack a portion of either head (lipid-III) or tail (“short” lipid-II) were studied in PG/PE membrane (see Methods).
“Short” analog of lipid-II does not cause formation of such prominent atoll-like pattern in the membrane (not shown) as lipid-II does. This is confirmed by analysis of size and lifetime distribution of hydrophobic clusters in the membrane (Fig. 5). On the contrary, lipid-III produces large and contrast pattern in PG/PE membrane (not shown), which is also evident form the cluster analysis (Fig. 5). Generally, this confirms the role of long and flexible lipid-II bacterioprenol tail in the disturbance of the nearby membrane.
Membrane behavior of lipid-II in the presence of other lipid-II molecules
In order to model more biologically relevant situation when lipid-II molecules have other lipid-II neighbors in the bacterial membrane (lipid-II membrane pool6), we have performed simulations of PG/PE membrane containing two lipid-II molecules (see Methods). In the course of 500 ns MD simulation we have not observed any tendency for these molecules to dimerize that corresponds well to the experimental data obtained in GUV, where lipid-II was found to be distributed homogeniously7. Thus, the distance between the lipid-II polar heads (measured as a distance between their PP groups) fluctuates around the initial value of 3.5 nm during the whole length of MD trajectory (Fig. 6B, inset). (One should keep in mind that two molecules in our setup could not dispart farther than ≈4 nm just because of the system size.) At the same time, terminal lipid-II tail segments (9th, 10th and 11th) are able to decrease the distance between them to 0.5–1 nm (Fig. 6A). Dynamics of tail segments depends on the distance of their chain from the lipid-II head. Spatial distances between closest to the head segments (1st and 2nd) evolve during MD in a correlated manner to those between PPs, while the correlation drops along the chain and becomes negative for the most terminal segments (Fig. 6B). In other words, if heads move towards each other, terminal segments display opposite character of motions and vice versa. As a result, the system of two lipid-II molecules displays oscillatory behavior on the equilibrium part of MD (Fig. 6B, inset), which helps them to maintain large enough distance, thus avoiding association in the membrane.
Behavior of two lipid-II molecules in PG/PE bilayer.
(A) Radial distribution functions (RDFs) for lipid-II bacterioprenol tail segments from two different molecules. Curves are colored according to the legend, where numbers mark the tail segments (starting from the closest one to PP) in the first and the second molecule, respectively. (B) Correlation of MD distances between lipid-II PP groups and their tail segments. Inset: MD evolution of distances between PP groups of two lipid-II molecules (black) and the last (11th) segments of their tails (red). Dashed vertical line depicts beginning of the “equilibrium” part of MD trajectory (150–500 ns), which was used for analysis of system with two lipid-II molecules. (C) MHP-map of PG/PE bilayer with the presence of two lipid-II molecules. Further details are available from the caption to Figure 3.
Discussion
Lipid-II normally works as a “shuttle carrier” of building blocks for the bacterial cell wall. At the same time, it possesses a conserved chemical structure and appears to be the target for many lantibiotics5,12. Our MD simulations of lipid-II in PG/PE membrane reveal that in the membrane-bound state many parts of lipid-II head group remain accessible to the solvent and thus can be targeted by various extracellular agents (Figs. 1 and 2). For instance, there is a body of evidence that lipid-II is recognized by many lantibiotics either via its pyrophosphate group (PP)5,10,11,12 (see Fig. 1A for lipid-II chemical structure) or via other head group parts, e.g. D-alanyl–D-alanine C-terminal dipeptide in a case of vancomycin5 or sugar residues in addition to PP in a case of nisin10, mersacidin14 and ramoplanin15.
Analysis of MD trajectory reveals that lipid-II is “anchored” at the water–lipid interface with its PP moiety and both head and tail groups are relatively flexible, especially the last one. The most structurally rigid part of the head is PP group with both sugar residues and first two residues of the pentapeptide. The peptide C-terminus is rather flexible and dynamically “shields” the rest of the head group from the solvent like an umbrella (Fig. 2). Maximal elevation of the C-terminus above the membrane surface is ≈2 nm, which is consistent with the data of supported-bilayer atomic force microscopy (AFM) experiment for lipid-II/dioleylphosphatidylcholine (DOPC) domains in the dipalmitoylphosphatidylcholine (DPPC) phase9. However, the mean “height” of the head group is much smaller (Supplementary Fig. S4 online), what is in agreement with reversible penetration of AFM tip into lipid-II while imaging at elevated force9.
Specific interactions between lipid-II and phospholipids modulate a particular membrane-bound conformation of lipid-II head group and the respective “membrane response”. In PG/PE membrane the following moieties of lipid-II molecule form extensive hydrogen-bonding networks with head groups of phospholipids: PP, N-acetylmuramic acid (“M-sugar”) and two of the pentapeptide residues — γ-D-glutamyl and lysyl (Fig. 1E). Analysis of lipid phosphorus atoms RDFs with respect to PP outlines its preferential interaction with POPE lipids (see Fig. 4 and video S3), which might contribute to unique structural and dynamic behavior of lipid-II in the simulated bacterial membrane as compared to the reference POPC bilayer.
According to the results of MD simulations, N-acetylglucosamine (“G-sugar”) represents the most solvent accessible part of lipid-II head group (Supplementary Fig. S1 online), although biochemical and NMR experiments show that it is not required for nisin and ramoplanin binding5,10,25. At the same time, “G-sugar” still might play a role in the interactions with mersacidin5. Less accessible PP group and “M-sugar” have been, however, considered as the major structural determinants of many lantibiotics binding12, given that pentapeptide seems to be not involved in binding of nisin25. Substantial embedding of these moieties at the water–lipid interface assumes possible interaction of PP-targeting lantibiotics with the membrane and the atomistic-level picture of this interaction is still highly required. The only structure of nisin complex with short tail lipid-II obtained by NMR spectroscopy in DMSO solution contains the so-called “pyrophosphate cage” spatial motif which has been proposed as a key element of lipid-II recognition by this lantibiotic11. However, this finding apparently requires refinement considering the real membrane-bound conformation of full-length lipid-II, taking into account that the membrane can be a part of the lantibiotics' target.
The part of lipid-II molecule that spans phospholipids' acyl chains — 11-mer bacterioprenol hydrophobic tail — features very unusual structure and dynamics. Chemical structure of the tail is typical for bipolar archaean lipids which pierce the whole membrane and have both polar termini fixed at the opposite surfaces of the bilayer26,27. Apart from these molecules, lipid-II tail does not have the second polar head that results in the unfixed tail terminus. Being so flexible, the tail of lipid-II explores the whole depth of hydrophobic slab in the simulated bacterial membrane mimic, where its terminus is travelling from one membrane surface to the opposite (Figs. 1B-D, 2A). Though, the most probable is “L-shaped” conformation of the tail (≈70% of MD time), when the terminus is located approximately in the middle of the hydrophobic core (Supplementary Fig. S3 online). Such a tail dynamics significantly affects the structure of the surrounding membrane and very likely contributes to interactions with lantibiotics.
Indeed, acyl chains of POPG or POPE lipids from the local vicinity of lipid-II molecule are prominently disordered according to the calculated Scdvalues (Fig. 3B) as compared to pure PG/PE bilayer or proximal areas of the lipid-II containing system (higher Scd values; Fig. 3A). The effect of local disordering of the lipid bilayer in the vicinity of lipid-II molecule has already been shown in previous MD simulation studies20, although no exhaustive picture of lipid-II dynamics and membrane response have been presented.
Disturbance of the membrane core obviously cannot leave the surface unaffected. Particularly, using the original mapping techniques of membrane surface properties (see Methods) we have found unique “amphiphilic pattern” that emerges around lipid-II molecule in the simulated bacterial membrane mimic as a consequence of aforementioned dynamic behavior of membrane-bound lipid-II. This pattern resembles an “atoll” with a horseshoe-like extended hydrophobic “terrain” that surrounds the hydrophilic “knob” (corresponding to the head group of lipid-II; Fig. 3D). The surface of lipid bilayers has been shown to possess mosaic distribution of hydrophobic and hydrophilic properties, which is however very dynamic28 and residing in a sub-nanometer scale29. The amphiphilic pattern around lipid-II is relatively extended (approx. 7 nm2) and long-living (300–400 ns; see Fig. 5B), what unambiguously discriminates it from the “basal” fluctuations of the lipid bilayer (Fig. 3C).
On this basis, the described amphiphilic pattern might serve as a unique binding platform, which can be recognized by lantibiotics. This phenomenon is described for the first time here, since it is hardly reachable for any experimental techniques and it has not been studied computationally in sufficient details. Importantly, truncation of either head or tail groups precludes formation of the “amphiphilic pattern” to different extent. Lack of the most of head group (in the case of lipid-III, Fig. 1A) leads to reduction of lifetime of hydrophobic clusters on the surrounding membrane surface but leaves the mean cluster size almost unaffected. This finding is consistent with the fact that nisin is still able to bind to lipid-III, although less efficiently25. At the same time, reduction of lipid-II tail (in a case of short tailed analog of lipid-II, Fig. 1A) seems to be more dramatic for the pattern formation: both lifetime and characteristic size of hydrophobic clusters decrease substantially (see Figs. 5 and S6).
The pattern created by lipid-II on the surface of simulated bacterial membrane is absent in the reference POPC bilayer (compare Fig. 3D with S5D). A possible explanation can be related to rather fluid and less “mosaic” structure of POPC membrane as compared to PG/PE mixture. In the latter case, more rigid water–lipid interface structure strengthened by extended H-bond and salt-bridges network30 promotes stabilization of lateral heterogeneities on the surface and contributes to their size and stability.
In such a manner, in PG/PE membrane head groups of neighboring PLs' (especially POPE lipids) are almost permanently attached to the lipid-II head group (in particular, PP) and remain bound for a long time (see Fig. 4 and video S3). At the same time, the tail of lipid-II disturbs acyl chains of these PLs and forces them to resurface and generate hydrophobic patches at the membrane surface (so-called “atoll”; see Fig. 3D). By contrast, in more fluid and less interconnected POPC lipid-II head group does not bind PLs' head groups so strongly and the energy of tail oscillations dissipates along with stirring of the neighboring areas of the membrane. As a result, no pronounced amphiphilic pattern is observed in POPC bilayer (Supplementary Fig. S5 online): hydrophobic heterogeneities are very small and short-living (Fig. 5 and Supplementary Fig. S6 online). As opposed to PG/PE membrane, lipid-II tail preferentially disintegrates hydrophobic clusters rather than promotes their formation.
As is evident, the choice of the lipid content may affect results of lipid-II studies. Although, many experimental9 and computational20 works were done with POPC or DOPC bilayers that differ from the native environment of lipid-II molecule. (At the same time, there is a study in POPG bilayer31.) We implicate that future investigations on lipid-II biochemistry and biophysics should be conducted with the use of membrane systems that mimic bacterial membrane more closely.
Although lipid-II is relatively fluid molecule in artificial bilayers preferring to localize in disordered lipid phase9 and distributed homogeneously in GUV7, it resides predominantly in specific parts of real bacterial membranes (pools)6 being surrounded by other lipid-II molecules. Two reasonable questions arise: are lipid-II molecules able to form oligomeric structures in the native-like environment and what will be the surface pattern structure in such a condition? According to results of MD simulations, two lipid-II molecules in the simulated bacterial membrane display no tendency for dimerization and only their tail terminal segments approach significantly to each other to be able to interact directly (Fig. 6A,B). This possible repulsion between the terminal tail groups might serve a mechanism of prevention of aggregation of lipid-II molecules in the bacterial membrane. Moreover, this effect allows a given lipid-II molecule to keep the same surface pattern in the presence of a neighbor, as it found for the single molecule (Fig. 3D and 6C) and form possible “recognition” site for the enzymatic machinery as well as lantibiotics. Experiments show that four lipid-II molecules may aggregate with eight nisin molecules to comprise a pore complex32. However, the stoichiometry 4:8 does not imply direct interaction between two lipid-II molecules and the aggregation itself may be peptide-driven.
Cell wall is one of the major vulnerable spots of bacteria and many antibiotics starting from penicillin act by inhibiting its synthesis. Variety of antibacterials, including those targeting resistant strains, is developed to attack bacterial envelope1. Lantibiotics act on presumably the most attackable site of cell wall synthesis — lipid-II — that is conserved chemically and unlikely to change its structure, apart from protein targets that easily evolve4,5,6. It has been demonstrated by biochemical and biophysical experiments that many lantibiotics directly interact with lipid-II and this interaction increases membrane affinity of these antimicrobial peptides up to three orders of magnitude19,33. One of the interesting biophysical questions here is the mechanism of intermolecular recognition, given that lipid-II is far from being abundant in the membrane (about few thousands molecules per cell) and antibiotics may be active in a very low concentration8.
It is reasonable to suppose that the binding mode of antibacterial agent to lipid-II incorporated into the bacterial membrane would differ from those obtained in structural studies of the isolated molecules in solution11. Although PP group and optionally other lipid-II's head fragments that are specifically recognized by lantibiotics18 still remain reachable from the outer membrane interior according to results of our simulations and AFM studies9, membrane surface surrounding lipid-II is very likely to contribute to the binding. We speculate that the “amphiphilic pattern” formed is a real lantibiotics' target in the bacterial membrane. The possible evidence for this is the fact that hydrophobic/hydrophilic properties of latibiotics' surface in the vicinity of lipid-II recognition site reflect the respective features of the “amphiphilic pattern”. Figure 7 illustrates this hypothesis by presenting hydrophobic/hydrophilic organization of “pyrophosphate cage” of nisin in complex with lipid-II11. It is clearly seen that the amphiphilic surface organization is featured not only in the membrane-bound lipid-II, but also in the corresponding part of nisin. Thus, as mentioned above, the real structure of this complex in the bacterial membrane environment might differ from the presented one, while the correspondence of physico-chemical properties of the ligand (lantibiotic) and the target (membrane with embedded lipid-II) should play an important role in the intermolecular recognition and in antibacterial action of these peptides.
“Amphiphilic pattern” on the surface of pyrophosphate cage motif of nisin suggests recognition mechanism.
The structure of nisin complex with lipid-II fragment is taken from PDB 1WCO11. Nisin is presented with semi-transparent surface colored by the Molecular hydrophobicity potential (calculated with PLATINUM43): blue color corresponds to hydrophilic areas, brown — to hydrophobic ones. Inside the surface nisin backbone is presented with sticks; lanthionine and methyllanthionine rings are colored yellow. In the front of nisin surface the fragment of lipid-II molecule is shown; probable recognition determinant is presented with sticks.
In this work we have performed an exhaustive computational study of lipid-II molecule in bacterial membrane — as we believe, both being the target of lantibiotic bactericidal action. For the first time, we give a detailed description of lipid-II dynamics that perturbs the membrane around itself and creates prerequisite for emergence of the “amphiphilic pattern” — a rather extended and long-living heterogeneity with increased hydrophobicity (formed from neighboring phospholipids' acyl chains). Along with the rise that is created by the “floating” head group of lipid-II over the membrane surface, this pattern may govern the initial stage of lipid-II recognition by lantibiotics. Apart from most of “traditional” targets, the one specific for nisin is formed by two interacting and mutually adopted partners — lipid-II and bacterial membrane. Each of them contributes its amphiphilicity and conformational lability into the creation of a unique 3D pattern, which can be potentially recognized by lantibiotics. To our knowledge, no similar dynamic multicomponent targets were described so far. Correct consideration of this mechanism may give new opportunities for studies on lantibiotics' action and design of novel compounds that selectively trap lipid-II and probably scavenge it, providing new armament against antibiotics-resistant bacterial strains.
Methods
Construction and molecular dynamics of systems
MD simulations of hydrated systems containing POPG/POPE (in this work referred to as PG/PE) or POPC phospholipid molecules arranged in bilayer structure were performed, with addition of different number and variants of lipid-II molecule(s) (Table 1). All simulations were carried out with the GROMACS 4.5.2 simulation suite34 using Gromos96 43a2x parameters set. The Ryckaet-Bellemans torsion potential was used for the lipid hydrocarbon chains35. Nonbonded interactions were described by the parameters from Berger et al.36. The force field parameters for POPC and POPE were taken from the website of Biocomputing laboratory at the University of Calgary (http://moose.bio.ucalgary.ca/index.php?page=Structures_and_Topologies) and that for POPG was elaborated based on parameters for the two first. Lipid-II topologies and initial systems coordinates that we have designed and used in this work are available at our website (http://model.nmr.ru/files/lipid-II_bilayers.zip).
Each simulation was performed using a unified protocol. First, PLs were arranged in a bilayer structure (with 144 molecules in each leaflet). In mixed systems, PLs of different types were distributed randomly in each leaflet. Second, lipid-II molecule(s) was (were) added to the bilayers; thereafter they were simulated for 50 ns and fully equilibrated. For this purpose, some PL molecules were removed from the bilayer. Third, the bilayer models were placed into rectangular boxes (typical size 8.0 × 8.0 × 1.1 nm3) and solvated with SPC37 water molecules and required number of Na+ ions to maintain electroneutrality. Prior to MD simulations, all systems were subjected to energy minimization (1000 conjugate gradients steps). Fourth and finally, each system was subsequently heated to 315 K in 60 ps and 500 ns of production MD runs were carried out for all considered systems, except for PG/PE with lipid-II, where 1000 ns trajectory was obtained.
Simulations were carried out with a time step of 2 fs and imposed 3D periodic boundary conditions, in the isothermal-isobaric (NPT) ensemble with semi-isotropic pressure of 1 bar and a constant temperature of 315 K. (This temperature was chosen to ensure that both lipid components are in fluid phase with some margin.) All components of the systems (water, phospholipids, lipid-II) were coupled separately to a temperature bath using Berendsen thermostat38 with a coupling constant of 0.1 ps. Van der Waals interactions were truncated using 1.2 nm spherical cut-off function. Electrostatic interactions were treated with the Particle Mesh Ewald (PME) algorithm39 (0.12 nm Fourier spacing). For further analysis (hydrophobicity and landscape mapping, as well as hydrophobic cluster analysis) trajectories were centered at lipid-II molecule using trjconv program from the Gromacs package34.
Analysis of MD trajectories
The computed 500/1000 ns MD trajectories were analyzed with the help of original software and modified utilities supplied with the GROMACS package.
Delineation and characterization of H-bonds in simulated membranes was performed using the g_hbond program from the Gromacs package34. Here, a hydrogen bond is defined according to the geometrical criteria, where the distance between the donor and acceptor is within 0.35 nm and the angle formed by donor, hydrogen and acceptor atoms is between 120° and 180°. We consistently analyzed H-bonds on time intervals of 50 ns and then chosen the most representative of them. If H-bond has existed for more than 1 ns during 50 ns, it was considered as stable and unstable otherwise.
Deuterium order parameters of acyl chains (SCD) were estimated using the g_order program from the Gromacs package34 as described in ref. 40. To analyze the spatial distribution of ordered/disordered phospholipids property, calculated order parameters were averaged over both acyl chains' carbon atoms for each phospholipid molecule separately (<SCD>). Phospholipids were considered as disordered if <SCD> was smaller than 0.1 and as ordered if <SCD> exceeded 0.25.
Lipid-II head group orientation was defined as an angle between the vector created by pyrophosphate (both phosphorus atoms and oxygen atom between them) with sugar and peptide moieties (one oxygen atom for each sugar and Cβ atom for each amino acid residue) and outward normal to the bilayer plane (Z = 0).
Pairwise radial distribution functions (RDFs) were calculated using g_rdf utility from the Gromacs package over the last 300 ns for each MD trajectory (main panel in Fig. 4).
2D-maps of hydrophobicity and landscape
To take advantage of clear and pictorial representation of bilayer surface and estimate lipid-II effect on the membrane (Figs. 2A,B, 4C, S2A,B and S3A,B) we calculated MD-averaged regular 2D maps that show distribution of hydrophobic/hydrophilic properties or landscape, respectively. This mapping was done as follows: 1. Each 100 ps of MD trajectory, coordinates of all bilayer atoms were saved to disk. 2. Connolly surface was created and a distribution of hydrophobic/hydrophilic properties was calculated in each surface point. These properties were approximated with the Molecular Hydrophobicity Potential (MHP) approach41,42 and calculated with PLATINUM software43. 3. An area of 7 × 7 nm2 of upper monolayer, centered on lipid-II molecule (if present), was used as a source for interpolation on a 70 × 70 Cartesian square grid with a step of 0.1 nm. Interpolated property was either MHP or average surface landscape (elevation or degradation with respect to the mean Z ≡ 0 nm). 4. Averaging was performed for each MD frame. 5. The averaged map was used for contour plotting and cluster analysis (see next paragraph). For steps 3–5, an in-house python script was developed that takes advantage of numpy, scipy and matplotlib modules.
Hydrophobic clusters analysis
MD snapshots from the last 470 ns of MD trajectories with time-separation of 100 ps were selected for analysis. MHP values on a regular grid (see previous paragraph) were used as a source data for the analysis. Points with MHP value ≥ 0.5 were considered as hydrophobic. The latter ones were clustered using two-scan connected component labeling algorithm44 in x-y plane for each frame. The largest cluster size in each frame was subjected to the analysis (Figs. 3 and S4). Clusters of size < 0.1 nm2 were neglected and did not take part in further calculations. The same algorithm was used to cluster hydrophobic points in 3-dimensional space: x-y-t, where t is MD time of the corresponding frame. This procedure gives the distribution of hydrophobic clusters lifetimes (Fig. 5B).
Analysis of interaction of two lipid-II molecules in a bilayer
Evolution of MD distances between PP groups as well as each bacterioprenol tail segment of two different lipid-II molecules was calculated using g_distance utility from the Gromacs package. RDFs for each tail segment in one lipid-II molecule with respect to those in the other one were obtained using g_rdf utility from the Gromacs suite. For MD evolution of distances between lipid-II PP groups and different tail segments Pearson correlation coefficients were also calculated. All aforementioned analyses were done for 150–500 ns of MD trajectory of two lipid-II molecules in PG/PE bilayer (Table 1).
References
Van Bambeke, F., Mingeot-Leclercq, M. P., Struelens, M. J. & Tulkens, P. M. The bacterial envelope as a target for novel anti-MRSA antibiotics. Trends Pharmacol Sci 29, 124–34 (2008).
Schneider, T. & Sahl, H. G. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol 300, 161–9 (2010).
Bierbaum, G. & Sahl, H. G. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10, 2–18 (2009).
Willey, J. M. & van der Donk, W. A. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61, 477–501 (2007).
Breukink, E. & de Kruijff, B. Lipid II as a target for antibiotics. Nat Rev Drug Discov 5, 321–32 (2006).
de Kruijff, B., van Dam, V. & Breukink, E. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fatty Acids 79, 117–21 (2008).
Hasper, H. E. et al. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–7 (2006).
Kramer, N. E. et al. Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol Lett 239, 157–61 (2004).
Ganchev, D. N., Hasper, H. E., Breukink, E. & de Kruijff, B. Size and orientation of the lipid II headgroup as revealed by AFM imaging. Biochemistry 45, 6195–202 (2006).
Bonev, B. B., Breukink, E., Swiezewska, E., De Kruijff, B. & Watts, A. Targeting extracellular pyrophosphates underpins the high selectivity of nisin. FASEB J 18, 1862–9 (2004).
Hsu, S. T. et al. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11, 963–7 (2004).
Hsu, S.-T., Breukink, E. & Kaptein, R. Structural Motifs of Lipid II-Binding Lantibiotics as a Blueprint for Novel Antibiotics. Anti-Infective Agents in Medicinal Chemistry 5, 245–254 (2006).
Milner-White, E. J., Nissink, J. W., Allen, F. H. & Duddy, W. J. Recurring main-chain anion-binding motifs in short polypeptides: nests. Acta Crystallogr D Biol Crystallogr 60, 1935–42 (2004).
Hsu, S. T. et al. NMR study of mersacidin and lipid II interaction in dodecylphosphocholine micelles. Conformational changes are a key to antimicrobial activity. J Biol Chem 278, 13110–7 (2003).
Hamburger, J. B. et al. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential Lipid II docking interface. Proc Natl Acad Sci U S A 106, 13759–64 (2009).
von Nussbaum, F. et al. Structure and total synthesis of lysobactin (katanosin B). Angew Chem Int Ed Engl 46, 2039–42 (2007).
Schneider, T. et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328, 1168–72 (2010).
Ross, A. C. & Vederas, J. C. Fundamental functionality: recent developments in understanding the structure-activity relationships of lantibiotic peptides. J Antibiot (Tokyo) 64, 27–34 (2011).
Breukink, E. et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–4 (1999).
Jia, Z., O'Mara, M. L., Zuegg, J., Cooper, M. A. & Mark, A. E. The effect of environment on the recognition and binding of vancomycin to native and resistant forms of lipid II. Biophys J 101, 2684–92 (2011).
Jia, Z., O'Mara, M. L., Zuegg, J., Cooper, M. A. & Mark, A. E. Vancomycin: ligand recognition, dimerization and super-complex formation. FEBS J (2013).
Dowhan, W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66, 199–232 (1997).
Morein, S., Andersson, A., Rilfors, L. & Lindblom, G. Wild-type Escherichia coli cells regulate the membrane lipid composition in a "window" between gel and non-lamellar structures. J Biol Chem 271, 6801–9 (1996).
Prossnigg, F., Hickel, A., Pabst, G. & Lohner, K. Packing behaviour of two predominant anionic phospholipids of bacterial cytoplasmic membranes. Biophys Chem 150, 129–35 (2010).
Muller, A., Ulm, H., Reder-Christ, K., Sahl, H. G. & Schneider, T. Interaction of type A lantibiotics with undecaprenol-bound cell envelope precursors. Microb Drug Resist 18, 261–70 (2012).
Itoh, Y. H., Sugai, A., Uda, I. & Itoh, T. The evolution of lipids. Adv Space Res 28, 719–24 (2001).
Gotoh, M. et al. Possible molecular evolution of biomembranes: from single-chain to double-chain lipids. Chem Biodivers 4, 837–48 (2007).
Polyansky, A. A., Volynsky, P. E., Arseniev, A. S. & Efremov, R. G. Adaptation of a membrane-active peptide to heterogeneous environment. II. The role of mosaic nature of the membrane surface. J Phys Chem B 113, 1120–6 (2009).
Pyrkova, D. V., Tarasova, N. K., Pyrkov, T. V., Krylov, N. A. & Efremov, R. G. Atomic-scale lateral heterogeneity and dynamics of two-component lipid bilayers composed of saturated and unsaturated phosphatidylcholines. Soft Matter 7, 2569 (2011).
Pyrkova, D. V. et al. Dynamic clustering of lipids in hydrated two-component membranes: results of computer modeling and putative biological impact. J Biomol Struct Dyn 31, 87–95 (2013).
Christ, K., Wiedemann, I., Bakowsky, U., Sahl, H. G. & Bendas, G. The role of lipid II in membrane binding of and pore formation by nisin analyzed by two combined biosensor techniques. Biochim Biophys Acta 1768, 694–704 (2007).
Hasper, H. E., de Kruijff, B. & Breukink, E. Assembly and stability of nisin-lipid II pores. Biochemistry 43, 11567–75 (2004).
Al-Kaddah, S. et al. Analysis of membrane interactions of antibiotic peptides using ITC and biosensor measurements. Biophys Chem 152, 145–52 (2010).
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced and stable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008).
Ryckaert, J. P. & Bellemans, A. Molecular dynamics of liquid n-butane near its boiling point. Chemical Physics Letters 30, 123–125 (1975).
Berger, O., Edholm, O. & Jahnig, F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure and constant temperature. Biophys J 72, 2002–13 (1997).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F. & Hermans, J. Interaction Models for water in relation to protein hydration. in Intermolecular Forces, ed. 331–342 (Reidel, 1981).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular-Dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577– 8592 (1995).
Lopez Cascales, J. J., Otero, T. F., Smith, B. D., Gonzalez, C. & Marquez, M. Model of an asymmetric DPPC/DPPS membrane: effect of asymmetry on the lipid properties. A molecular dynamics simulation study. J Phys Chem B 110, 2358–63 (2006).
Efremov, R. G. et al. Molecular lipophilicity in protein modeling and drug design. Curr Med Chem 14, 393–415 (2007).
Efremov, R. G., Gulyaev, D. I., Vergoten, G. & Modyanov, N. N. Application of three-dimensional molecular hydrophobicity potential to the analysis of spatial organization of membrane domains in proteins: I. Hydrophobic properties of transmembrane segments of Na+, K(+)-ATPase. J Protein Chem 11, 665–75 (1992).
Pyrkov, T. V., Chugunov, A. O., Krylov, N. A., Nolde, D. E. & Efremov, R. G. PLATINUM: a web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics 25, 1201–2 (2009).
He, L., Chao, Y. & Suzuki, K. A run-based two-scan labeling algorithm. IEEE Trans Image Process 17, 749–56 (2008).
Acknowledgements
This work was supported by the Ministry of Education and Science of the Russian Federation (contract no. 07.514.11.4127), by the Russian Foundation for Basic Research and by the RAS Programmes “Molecular and cellular biology” and “Basic fundamental research for nanotechnologies and nanomaterials”. A.O.C. is recipient of the Stipend of the President of the Russian Federation. Access to computational facilities of the Joint Supercomputer Center RAS (Moscow) and Moscow Institute of Physics and Technology is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
A.O.C., A.A.P., V.M.P. and R.G.E. designed the research, A.O.C., D.V.P., D.E.N. and A.A.P. performed calculations and data analysis, D.E.N. set up computer cluster, A.O.C. and A.A.P. prepared figures, A.O.C., A.A.P. and R.G.E. wrote the manuscript, all authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary video S1
Supplementary Information
Supplementary video S2
Supplementary Information
Supplementary video S3
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Chugunov, A., Pyrkova, D., Nolde, D. et al. Lipid-II forms potential “landing terrain” for lantibiotics in simulated bacterial membrane. Sci Rep 3, 1678 (2013). https://doi.org/10.1038/srep01678
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01678
This article is cited by
-
Deciphering the Limitations and Antibacterial Mechanism of Cruzioseptins
International Journal of Peptide Research and Therapeutics (2022)
-
The Antibiotic Peptide Daptomycin Functions by Reorganizing the Membrane
The Journal of Membrane Biology (2021)
-
Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids
Nature Communications (2020)
-
Exploring metabolic adaptation of Streptococcus pneumoniae to antibiotics
The Journal of Antibiotics (2020)
-
Environmental and dynamic effects explain how nisin captures membrane-bound lipid II
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.