Abstract
Sex determination cascade in insects terminates with the production of sex-specific protein, Doublesex (Dsx). We identified the dsx homolog (Tcdsx) in Tribolium castaneum. The pre-mRNA of Tcdsx is sex-specifically spliced into three female (Tcdsxf1, Tcdsxf2 and Tcdsxf3) and one male-specific (Tcdsxm) isoforms. Cis-regulatory elements potentially involved in sex-specific splicing of the Tcdsx pre-mRNA were identified in the female-specific exon and the adjoining intronic sequences. All the three female-specific TcDsx proteins share common OD1 and OD2 domains and differ in their C-terminal sequences. Knockdown of Tcdsx resulted in a reduction in the oocyte development, egg production and hatching of eggs laid. Several genes, including those coding for Vitellogenins and Vitellogenin receptors were identified as targets of TcDsx. RNAi experiments showed an isoform-specific targeting of identified target genes by TcDsx as knockdown in the expression of Tcdsx isoforms individually or in combinations resulted in differential effects on the expression of target genes.
Similar content being viewed by others
Introduction
Surprising diversity is shown by the organisms in their sex determination mechanisms1,2. Insects are no exception; different strategies are utilized by insects belonging to different orders to determine their sex during the early embryonic development3,4,5,6. The diversity in sex-determination mechanism lies in the presence of different upstream signals which ultimately work through the DM (Doublesex-Mab3) domain containing transcription factors3,7. The DM domain containing transcription factors are the best characterized sexual differentiation proteins7,8 that are conserved among insects (doublesex- dsx)9, worms (male abnormal3- mab3)10 and vertebrates (doublesex and mab3 related transcription factor- dmrt)11. The dsx is the bottom most gene of the sex determination cascade in Drosophila melanogaster12,13. The pre-mRNA of dsx is sex-specifically spliced to produce one female- and one male-specific isoforms in turn generating one female (DsxF) and one male (DsxM) specific Dsx proteins, respectively. Sex-specific Dsx proteins share common DNA binding (DM or OD1) domain14 but differ within their oligomerization domain (OD2)15. Due to this difference, sex-specific Dsx proteins have antagonistic effects on the regulation of their target genes involved in various aspects of sex differentiation13,16. Since the discovery of dsx in D. melanogaster, its homologues have been identified in several insect species belonging to orders Diptera17,18,19,20,21, Hymenoptera22,23 and Lepidoptera24,25,26. In most of the insect species studied, the pre-mRNA of dsx was found to be sex-specifically spliced to produce one female- and one male-specific RNAs. However, the dsx pre-mRNAs of Musca domestica27 and Apis mellifera22 are spliced to produce more than two splice variants. On the basis of open reading frame (ORF), one male- and one female-specific Dsx proteins are predicted. In Aedes aegypti17, Bombyx mori24,28 and other wild silkmoths25 more than one dsxf transcripts have been identified which ultimately may generate more than one female-specific Dsx proteins. RNAi mediated knockdown studies showed the requirement of both the DsxF proteins in the female sexual differentiation of silkmoths25. Several indirect targets of Dsx have been predicted in D. melanogaster29,30,31. Recently, several direct targets of Dsx have been predicted, in D. melanogaster, based on the Dsx binding sites present in the promoter and intergenic region of these genes32. Very few genes have been functionally demonstrated to be the direct Dsx targets, among them vitellogenin (vg) is the best example16,33. The regulation in the expression of genes coding for Vg, Pheromone binding protein and hexamerin by Dsx has been shown in the lepidopteran insects25,34,35. Recent studies showed that bric-a-bric (bab)36, fad237 and wingless (wg)38 genes are also regulated by Dsx in D. melanogaster.
In spite of the fact that the insect order Coleoptera contains one fourth of all species described and includes many major pests of crop plants39, nothing is known about the dsx and its targets in this group of insects. We identified and characterized the dsx homologue (Tcdsx) in coleopteran model insect, the red flour beetle, Tribolium castaneum. The pre-mRNA of Tcdsx is sex-specifically spliced to produce three female (Tcdsxf1, Tcdsxf2 and Tcdsxf3) and one male-specific (Tcdsxm) isoforms. All the three female-specific Tcdsx isoforms are generated as a result of alternative splicing within the female-specific exon (exon3). Interestingly, putative cis-regulatory elements were found in the female-specific Tcdsx exon and the adjoining intron sequences suggesting their possible involvement in the sex-specific splicing of Tcdsx pre-mRNA. We found several TcDsx target genes in T. castaneum by comparing the expression of previously identified female-specific genes in the control and Tcdsx RNAi insects. Knockdown in the expression of Tcdsx gene in an isoform-specific manner resulted in differential expression of identified target genes suggesting an isoform-specific regulation of target genes. The data included here confirm the evolutionary conserved role of dsx in insect sexual differentiation.
Results
Identification and characterization of T. castaneum doublesex
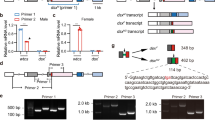
In order to identify dsx homolog (Tcdsx) in T. castaneum, blast (tblastn) searches were performed in the Beetlebase (http://beetlebase.org/) and in the NCBI (http://www.ncbi.nlm.nih.gov/) using known Dsx protein sequences as a query. A single sequence (ORF) of 969bp in length and annotated as “Tribolium castaneum similar to BmDSX-F (LOC660453)” was identified. Forward and reverse primers were designed based on this sequence (LOC660453). Three fragments were amplified when cDNA made using RNA isolated from females was used as a template in RT-PCR. Whereas, only one fragment was amplified when cDNA made using RNA isolated from male was used as a template (data not shown). Sequencing and analysis of sequence of these fragments showed that ‘LOC660453’ is a male-specific isoform (Tcdsxm) of Tcdsx. The three female isoforms identified are named as Tcdsxf1, Tcdsxf2 and Tcdsxf3. UTR sequences were obtained by sequencing 3′ and 5′ RACE PCR products which were confirmed by aligning them with the Tcdsx genomic sequences (AAJJ01001880.1 and AAJJ01000060.1). Further, full length Tcdsx splice variants were amplified by RT-PCR using sex-specific cDNA and primers specific to the ends of Tcdsx (Fig. 1A); three female-specific amplicons of 2264bp (Tcdsxf1), 2428bp (Tcdsxf2) and 2186bp (Tcdsxf3) and one male-specific amplicon of 1788bp (Tcdsxm) were obtained (Fig. 1B). Because, the difference between Tcdsxf1 and Tcdsxf3 is only 78bp, they migrate closely in the gel (Fig. 1B). The PCR fragments were cloned and sequenced and analysis of sequences confirmed the presence of two products in Tcdsxf1/f3 bands. Further, to show the presence of three female-specific Tcdsx splice forms, RT-PCR was performed using internal primers and sex-specific cDNAs; three female- and one male-specific amplicons were amplified (Fig. 1C). The conceptual translation of ORFs of these sex-specific isoforms showed the presence of DM and OD domains confirming the existence of three female- and one male-specific Tcdsx isoforms. Full length cDNA sequences and the deduced amino acid (aa) sequences of Tcdsx have been submitted to GenBank (accession no. for Tcdsxm, Tcdsxf1, Tcdsxf2 and Tcdsxf3 are JQ857098, JQ857099, JQ857100 and JQ857101, respectively).
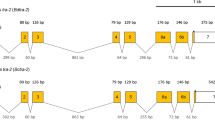
(A) Schematic representation of isoforms of Tcdsx pre-mRNA, showing the primer positions and regions used for preparation of dsRNA. Boxes show exons and lines show introns. The sizes (bp) of different exons are shown within the exons. Blue colored regions represent the ORF whereas the orange colored regions represent UTRs. Four different splice variants of Tcdsx pre-mRNA, three female- (Tcdsxf1, Tcdsxf2 and Tcdsxf3) and one male-specific (Tcdsxm), are produced. A(n) show the polyadenylation site. Vertical lines represent start and stop codon sites. Vertical arrows show the domains of the TcDsx proteins whereas horizontal arrows show primer positions. Green horizontal lines show regions corresponding to dsRNA; ds-dsxC (common) = 354 bp, ds-dsxf1+f2 = 78 bp and ds-dsxf2 = 120 bp. Primers F1 and R1 were used to amplify full length Tcdsx transcripts and primer qRTCF was used with either qRTf1R, qRTf2R or qRTf3R in qPCR for the quantification of specific Tcdsxf transcripts. The sequences of all the primers mentioned here are given in supplementary Table 2. (B) Gel picture showing three bands (Tcdsxf1, Tcdsxf2 and Tcdsxf3) in females and one band in male (Tcdsxm) as a result of RT-PCR using sex-specific cDNA as template and primers (F1 and R1) specific to ends of Tcdsx (Fig. 1A). M represents DNA size marker. C) Gel picture showing three bands (Tcdsxf1, Tcdsxf2 and Tcdsxf3) in females and one band in male (Tcdsxm) as a result of RT-PCR using sex-specific cDNA as template and internal primers (F2 and R2) spanning the alternatively spliced region of Tcdsx (Fig. 1A). Same primers (F2 and R2) were used for analyzing the splicing status of Tcdsx in previous paper55. M represents DNA size marker.
Genomic organization of Tcdsx and proteins encoded by Tcdsx isoforms
The Tcdsx transcript sequences span 8503bp region in AAJJ01001880.1 and 23138bp region in AAJJ01000060.1 genomic contigs (Fig. 1A). Exon-intron boundaries were assigned based on the alignment of Tcdsx cDNA sequences with the corresponding genomic DNA sequences (AAJJ01001880.1 and AAJJ01000060.1). Tcdsx gene harbors 6 exons and 5 introns; except for exon 3 which is female-specific, all others are common to both male and female (Fig. 1A). Generation of three female-specific transcripts is due to alternative splicing within female-specific exon (exon 3). The deduced amino acid sequences of three TcDsxF proteins differ with each other only at their C-terminal ends. First two exons, common to all the female- and the male-specific transcripts, code for 197aa common N-terminus region of TcDsx proteins. Due to the alternative splicing within the third exon 55, 30 and 29 aa are added to TcDsxF1, TcDsxF2 and TcDsxF3 respectively (Fig. 2A and Fig. S1). Multiple sequence alignment of sex-specific TcDsx proteins with the Dsx proteins from other insects (D. melanogaster, A. gambiae and B. mori) showed a high degree of sequence conservation in the N-terminal region containing DM/OD1 domain and in the common region containing the OD2 domain (Fig. 2A). Interestingly, amino acids residues (C, H, H, C, C and R) within the DM/OD1 domain shown to be essential for DNA binding activity in D. melanogaster14,41 were found to be 100% conserved in TcDsx proteins (Fig. 2A). Consistent with this, very high sequence conservation was also found in the C-terminal regions of the DsxF proteins (Fig. 2B). Very little similarity was observed in the male-specific regions of Dsx proteins (Fig. 2C). Unlike Dsx proteins identified from other insects, the male-specific TcDsx protein (TcDsxm) contains two OD2 domains (OD2a and OD2b). Whereas, female-specific TcDsx proteins contain only one OD domain (OD2a). TcDsxM and TcDsxF are identical at their N-terminus until the OD2a domain of TcDsxM (OD2a is truncated in males but complete in females) but differ at their C-terminal region; OD2b is specific to TcDsxM (Supplementary Fig. 1S). Supplementary Table 1 summarizes the length of mRNA, protein and number of exons present in different Tcdsx isoforms.
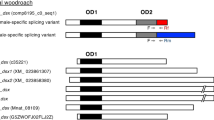
Deduced amino acid (aa) sequences of Dsx proteins from T. castaneum (TcDsx), D. melanogaster (DmDsx), A. gambiae (AgDsx) and B. mori (BmDsx) were aligned.
The Dsx sequences are divided into (A) region common to DsxM and DsxF proteins (B) Female-specific region which is known to be conserved and C) Male-specific region. The DNA binding domain (DM/OD1) is shown in blue color whereas oligomerization domain (OD2) is shown in orange color. Arrows indicate the conserved amino acid residues (in red) shown to be essential for DNA binding activity of Dsx in D. melanogaster. The three TcDsxF proteins differ from each other at their C-terminus (B). Additional OD2 domain in TcDsxM is shown in orange color (C). Stars (*) represent 100% identity of aa among all proteins.
Putative splicing regulatory elements in Tcdsx
In-silco analysis of splice sites (donor/acceptor sequences) of Tcdsx showed the presence of GT at all the splice donor sites and AG at the intron junctions between exons 2, 4 and 5 (Table 1). The splice acceptor site preceding the exon 3 also contains AG but the additional introns generated inTcdsxf1 and Tcdsxf3 (see Fig. 1) contain CT at their splice acceptor sites. The splice acceptor site in the intron preceding exon 6 contains TC rather than AG (Table 1). We did not find any repeats of putative dipteran Tra/Tra2 binding sequence in the genomic sequence of Tcdsx; the female-specific exon of dsx in dipteran insects contain imperfect repeat of 13nt (RE-Repeat Element)17,18,19,42. Interestingly, three repeats (A/CGAAGAAA/G) matching the putative dsx RE sequence of A. mellifera and Nasonia vitripennis43 were found exclusively in the intron downstream to the female-specific exon (exon3); these repeats are clustered within 552bp region. A Purine Rich Stretch (PRE- GAAGAAGTAGAGAA) was also identified exclusively in the 3′ region of exon 3 (Supplementary Fig. 2S). Clustering of the splicing regulatory sequences in the female-specific exon and the adjoining intron sequences suggests their possible involvement in the sex-specific splicing of the Tcdsx pre-mRNA.
RNAi studies on Tcdsx
T. castaneum is an excellent coleopteran model insect owing to the fact that RNAi woks efficiently in this beetle44. dsRNA 1) targeting common region of Tcdsx which could silence all the female- and male-specific transcripts, 2) targeting two female-specific transcripts, Tcdsxf1 and Tcdsxf2 together and 3) targeting only one female-specific transcript, Tcdsxf2 (Figure 1A), were synthesized and injected into newly eclosed sex-separated pupae of T. castaneum. Total RNA isolated from 5 day-old adult's eclosed from injected pupae was used to quantify Tcdsx mRNA levels. Injections of dsRNA caused an efficient knockdown in the expression of targeted Tcdsx isoforms (Supplementary Fig. 3S). The phenotypes observed in Tcdsx knockdown insects are described below.
Effect of Tcdsx RNAi on oocyte development
In T. castaneum, oocyte development takes place during the first few days post adult emergence (PAE)45,46. Ovaries were dissected from 5th day PAE females, eclosed from pupae injected with Tcdsx or malE dsRNA, stained and observed under a microscope. When compared to the oocytes in the control beetles injected with malE dsRNA (Fig. 3A), there was a significant reduction in the size of oocytes in the females injected with dsRNA targeting all the three Tcdsx transcripts together or Tcdsxf1 plus Tcdsxf2 or Tcdsxf2 alone (Fig. 3B, 3C and 3D, respectively). The primary oocytes in control insects matured to stages 6-7 [see Parthasarathy et al46 for description of oocyte stages] whereas, the primary oocytes in Tcdsx dsRNA injected insects were arrested at stages 1–4. Injection of Tcdsx dsRNA (targeting common region) in males affected testis development; the lobes in the testis of Tcdsx RNAi insects are smaller than that of control insects injected with malE dsRNA (Fig. 3E and 3F). These data suggest the requirement of TcDsx proteins for the development of oocytes and testis.
Tcdsx RNAi affects development of ovary and testis.
Young (0 day) sex-separated pupae were injected with dsRNA targeting all isoforms of TcdsxC (f1+f2+f3), Tcdsxf1+ Tcdsxf2 (f1+f2), Tcdsxf2 alone or malE. Ovaries or testis were dissected on 5th day PAE, stained with acridine orange and photographed at 4X (ovary) or 10X magnification (testis) using fluorescence microscope.
Effect of Tcdsx RNAi on egg production and embryogenesis
We also determined the effect of Tcdsx knockdown on egg production and hatching of eggs laid, by crossing female and male adult beetles eclosed from pupa injected either with Tcdsx or malE dsRNA. Crosses were done in four different combinations; Tcdsx RNAi females with Tcdsx RNAi males, Tcdsx RNAi females with malE RNAi males, malE RNAi females with Tcdsx RNAi males and malE RNAi females with malE RNAi males. No eggs were produced by the Tcdsx RNAi females mated with Tcdsx RNAi or control (malE RNAi) males in all the Tcdsx dsRNA injected beetles (Table 2). Females developed from uninjected pupae or pupae injected with malE dsRNA and mated with males developed from uninjected pupae or pupae injected with malE dsRNA laid the same number of eggs. Very few eggs were laid by the females mated with Tcdsx RNAi males but these eggs hatched and developed into larvae (Table 2).
Effect of Tcdsx RNAi on vg mRNA levels
The gene coding for Vitellogenin (Vg) has been characterized as a target for Dsx protein in D. melanogaster47. Previous studies have shown that the basal levels of vg gene expression is up regulated by the female form of Dsx protein (DsxF) and down regulated by the male form of Dsx protein (DsxM)16,33,47,48. In T. castaneum, two genes [vg1 (Glean-13602) and vg2 (Glean-10839)] coding for Vg have been identified and both the genes show identical patterns of expression49. Both vg1 and vg2 mRNA levels were quantified in Tcdsx knockdown and control insects. dsRNA injections, targeting common region of Tcdsx, lead to a significant (p<0.05) reduction in both vg1 and vg2 mRNA levels in females (Fig. 4A). Knockdown of Tcdsxf1 and Tcdsxf2 together, also lead to a significant reduction in both vg1 and vg2 mRNA levels in females. No significant reduction in both vg1 and vg2 mRNA levels were observed in females injected with Tcdsxf2 dsRNA, as compared to the control females injected with malE dsRNA (Fig. 4A). Both vg1 and vg2 mRNA levels in male Tcdsx RNAi insects were significantly higher (p<0.05) as compared to their levels in control males injected with malE dsRNA (Fig. 4B).
Relative expressions of female-specific genes in Tcdsx or malE dsRNA injected beetles.
Young (0 day) sex-separated pupae were injected with dsRNA targeting all isoforms of TcdsxC (f1+f2+f3), Tcdsxf1+ Tcdsxf2 (f1+f2), Tcdsxf2 alone or malE. Total RNA was isolated on 5th day PAE and the mRNA levels of selected genes were quantified by qRT-PCR. Mean+S.D (n = 3) relative mRNA levels are shown. Asterisks show treatments that are significantly different from control (p<0.05). (A) TcDsx target genes that are down regulated in females injected with dsRNA targeting either common region of Tcdsx or the region specific to Tcdsxf1 and Tcdsxf2 (f1+f2) together. (B) Female-specific genes that significantly increased in Tcdsx RNAi males. (C) TcDsx target genes that decrease only when dsRNA targeting the common region of Tcdsx was injected into females. The expressions of these genes are unaffected in females injected with dsRNA targeting Tcdsxf1 and Tcdsxf2 together. All these genes are unaffected by the injection of dsRNA targeting Tcdsxf2 alone.
Identification of TcDsx target genes
Microarray studies using RNA isolated from sex-separated adults of T. castaneum insects identified several genes expressed in a female-specific manner (data not shown). We selected 30 female-specific genes based on their minimum of 5-fold expression difference between female and male and tested the possibility of these genes as targets of TcDsx protein. cDNA made from RNA isolated from Tcdsx RNAi (knockdown of a single or a combination of Tcdsx isoforms) and control beetles and primers (supplementary Table 2) specific to the selected female-specific genes were used in the qRT-PCR analysis to quantify mRNA levels. Out of the 30 genes tested, 12 genes (including two vg and two vg receptor genes) showed a reduction in their mRNA levels in Tcdsx RNAi females compared to that in control females suggesting that these genes may be regulated by TcDsx (Fig. 4A and 4B).
Differences in the expression of these genes in beetles injected with dsRNA targeting different isoforms of Tcdsx were also observed. On this basis, these genes were categorized into two groups; I) genes, whose expression is down regulated when the dsRNAs targeting the common region or the region specific to both, Tcdsxf1 and Tcdsxf2 transcripts were injected (Fig. 4A) and II) genes, whose expression is down regulated when dsRNA targeting the common region of Tcdsx is injected (Fig. 4C). These data suggest the possibility of regulation of different genes by TcDsxF proteins in an isoform-specific manner. Interestingly, there is no difference in the expression of any of these target genes in the females injected with dsRNA targeting only Tcdsxf2 isoform (Fig. 4A and 5C) suggesting a possible functional redundancy of TcDsxF2 withTcDsxF1 and TcDsxF3. We did not find any significant difference in the down regulation of these genes when dsRNA targeting the common region was injected compared to when dsRNAs specific to Tcdsxf1 and Tcdsxf2 transcripts were injected (data not shown). Injection of dsRNA targeting common region of dsx into males caused a significant increase in mRNA levels of both vg genes and G07776 (Fig. 4C) suggesting that TcDsxM suppresses expression of these three genes in males. The expression of other nine genes tested was not affected in Tcdsx RNAi male beetles.
(A) Schematic representation of genes showing the presence of putative Dsx binding site sequences in the putative promoter and intergenic region of eight (G03183, G08596- Vg receptor, G04042- Vg receptor, G15076- Proteoglycan 4, G13602- Vg1, G10839- Vg2, G14653- Lysosomal acid lipase and G07776) of the TcDsx target genes. (B) Weblogo, showing the nucleotide identity of the putative TcDsx binding site sequences in the identified TcDsx target genes. The height of each stack indicates the consensus sequences at that position (in bits) whereas the height of nucleotides represents the relative frequency of each nucleotide at that position.
Direct Dsx target genes in D. melanogaster are known to harbor a 13bp palindromic sequence to which Dsx binds15,32. Computational analysis of the whole genome sequence of different insect species besides dipterans suggest an evolutionary conservation and enrichment of Dsx-binding sequence in their genome32. In order to search for the Dsx binding site(s) and their location in the identified TcDsx targets we scanned the genomic region corresponding to each gene (the region of genomic DNA containing gene + 3 kb upstream to their ATG site) for the presence of sequence(s) similar to Dsx binding sites identified in D. melanogaster. Interestingly, we noticed the presence of 13 bp consensus sequence(s) in eight (G03183, G08596- Vg receptor, G04042- Vg receptor, G15076- Proteoglycan 4, G13602- Vg1, G10839- Vg2, G14653- Lysosomal acid lipase and G07776) of these genes (Fig. 5A and 5B) suggesting that these genes may be the direct targets of TcDsx proteins. These Dsx binding sequences are composed of two half-sites around a central (A/T) base pair, the core (7 bp) region of which was found to be palindromic and highly conserved as reported for Dsx binding sites in D. melanogaster32. These putative TcDsx binding sites are located in the promoter or intergenic regions (except in G13602 where one of the putative Dsx binding sequences is also found in exonic region). Table 3 summarizes the identified Tcdsx target genes, corresponding genomic contig, tissue of expression and their putative functions.
Discussion
One of the final steps of sex determination cascade in insects is the production of female- or male-specific Dsx proteins4,9,50. Production of one female- and one male-specific dsx isoforms (in turn one female- and one male-specific Dsx protein) is considered to be the trade mark feature of dsx in the insect species studied so far. The existence of one female- and one male-specific Dsx proteins was unchallenged until the discovery of multiple female-specific dsx pre-mRNAs, differing in their ORFs, in silkmoths25,28 and yellow fever mosquito17. Identification of multiple isoforms of Tcdsx and the results from the RNAi-mediated knockdown of these splice variants in different combinations supports the requirement of more than one DsxF proteins in the female sexual differentiation. All the three female-specific Tcdsx transcripts are expressed during larval (final instar larvae), pupal and adult stages (data not shown). The expression of four genes (G03183; G08596,Vg receptor; G04042,Vg receptor and G15076,Proteoglycan 4) was reduced in females injected with dsRNA targeting the common region of Tcdsx transcripts but unaffected by the injection of dsRNA targeting Tcdsxf1 plus Tcdsxf2 transcripts (Fig. 4C). There could be two possibilities for the observed difference in both the cases; 1) the knockdown efficiency of Tcdsxf1 and Tcdsxf2 transcripts are different in both the cases since the dsRNA targeting different regions were synthesized and they varied in their lengths (Fig. 1A) and/or 2) these genes are regulated by TcDsxF3 protein alone. Quantitative analysis of the levels of Tcdsxf1 and Tcdsxf2 transcripts in the insects injected with dsRNA targeting either common region or Tcdsxf1 and Tcdsxf2 transcripts specifically showed almost equal amount of reduction of these transcripts in both the cases (Supplementary Fig. 3S). Hence, we conclude that TcDsxF3 protein is the sole regulator of these genes. Other group of genes showed highly reduced expression in females injected with dsRNA targeting common region or Tcdsxf1 plus Tcdsxf2; the expression of these genes was unaffected by injection of dsRNA specific to Tcdsxf2 alone. At present, it is difficult to conclude whether the expression of these genes is regulated by TcDsxF1 or TcDsxF3 or by both these proteins. Either we did not identify any TcDsxF2 target gene or function of TcDsxF2 is redundant with other TcDsxF isoforms. Presence of underdeveloped oocytes in the females injected with dsRNA targeting Tcdsxf2 alone suggests a role for Tcdsxf2 in oocyte development and hence most likely TcDsxF2 target genes were not identified in our analysis. Further, the presence of putative TcDsx binding sequences in some of the TcDsx target genes (Fig. 5A and 5B) suggest them to be the direct targets of TcDsx. Other genes (G07775, G05384, G07186 and G06658) in which no conserved Dsx binding sites were identified are most likely indirect targets of TcDsx as Dsx in D. melanogaster is known to regulate many transcription factors which in turn regulate the expression of genes51.
The female-specific exon 4 of D. melanogaster dsx contains a stretch of purine nucleotides which makes the preceding 3′ splice site a weak splice acceptor and therefore overlooked by the spliceosomal machinery in males, resulting in the skipping of exon 452. In females, Transformer (Tra), a female-specific protein along with Tra2 (a non sex-specific protein) and with other splicing regulatory proteins binds to the dsx repeat elements (dsxRE) making the 3′splice site stronger resulting in the incorporation of exon 4 in female-specific dsx isoform52,53,54. We recently characterized the homolog of tra (Tctra) in T. castaneum and showed its requirement for the female-specific splicing of Tcdsx55. The Tctra pre-mRNA, in males, splices in a manner which retains male-specific exons with several in-frame stop codons in it, resulting in the production of non-functional protein55. Several cis-regulatory sequences were found to be located exclusively in the female-specific exon and its adjoining intron of Tcdsx. Together with the observation of skipping of exon 3 in males and presence of a stretch (14 bp) of purine rich sequence (GAAGAAGTAGAGAA), in the 3′ end of female-specific exon suggest this as putative Purine Rich Element (PRE) of Tcdsx which most likely makes the 3′ splice site preceding exon 3 as a weak splicing acceptor site. Besides, clustering of putative cis-regulatory elements (putative Tra/Tra2 binding sequences and Tra2-ISS elements) was found in the female-specific exon (exon 3) and the adjacent intron sequences. Identification of these putative cis-regulatory elements together with the previous data55 suggest the interaction of TcTra/Tra2 protein complex and other splicing regulatory proteins with the cis-regulatory elements present in Tcdsx, in females and regulate female-specific splicing (retention of exon3) of Tcdsx pre-mRNA. However, the mechanisms of regulation of alternative splicing within the exon 3 and generation of three female-specific Tcdsx isoforms remains unknown. Presence of weak splice acceptor site (CT) within exon 3 might be playing a role in alternative splicing of female-specific exons. Similar kind of alternative splicing within the female-specific exons of dsx leads to the generation of two female-specific splice variants with different ORFs in lepidopteran insects25,28. Studies on differential expression of target genes in beetles injected with dsRNA targeting one or more Tcdsx isoforms and effect of knockdown in the expression of specific Tcdsx isoforms on oocyte development provided some initial clues on isoform-specific functions of Tcdsx. However, further studies are needed to clarify isoform-specific functions of dsx.
Methods
Tribolium castaneum strain, RNA isolation and RT-PCR
The GA-1 strain of the red flour beetle, Tribolium castaneum was used in all the studies. Organic whole wheat flour containing 10% yeast was used for rearing the beetles. Pupa and adults were sexed based on the presence of sex-specific structures. RNA was isolated using Trizol method (Invitrogen Corporation, USA). DNAse treated total RNA was denatured at 75°C for 5 min and immediately chilled on ice. First strand cDNA was synthesized with MMLV reverse transcriptase (Invitrogen, USA) using 17-mers polyT primer. Initial denaturation at 94°C for 2 min, 32 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 2 min and the final extension at 72°C for 10 min were used to perform PCR reactions.
3′ and 5′ RACE PCR
3′ and 5′ RACE reactions were performed using SMARTer RACE cDNA Amplification Kit (Clontech, USA) according to manufacturer's instruction, using adapter- and gene-specific primers. The primary and nested gene-specific primers used for 3′ RACE PCR of Tcdsx are 3′GSP1_dsx (5′-GATCAAGACGAGGCGTGGAGGCAGAT-3) and 3′GSP2_dsx (5′-CGCCGTCGAAGCAGCAAGGTACAC-3′) The primary and nested gene-specific primers used for 5′ RACE PCR are 5′GSP1_dsx (5′-CTCGGTGGTCAGCCGGCACTTCT-3′) and 5′GSP2_dsx (5′-CTTGTGGCCTTTGAGGGCGATCTTGA-3′).
Drop down PCR reaction with conditions, initial denaturation at 94°C for 2 min, 5 cycles of 94°C for 30 s, 72°C for 2 min, 5 cycles of 94°C for 30 s, 70°C for 2 min, 25 cycles of 94°C for 30 s, 68°C for 2 min, 72°C for 2 min and a final extension at 72°C for 10 min were performed on Biorad master cycler. Amplicons of different sizes were gel-eluted, sequenced and the 5′ and 3′ Tcdsx sequences were selected based on the overlapping regions of known sequence of Tcdsx. Sequences were further confirmed by RT-PCR using primers specific to the known sequence and the sequences obtained through RACE experiments.
Sequence analysis
Exons and introns, of the Tcdsx were identified by aligning sequences of RT-PCR products with their corresponding genomic DNA sequences obtained from the Beetlebase (http://beetlebase.org/) and the NCBI (http://www.ncbi.nlm.nih.gov/). Exon-intron boundaries were confirmed by aligning the sequences through Spidey program (http://www.ncbi.nlm.nih.gov/spidey/).
Double stranded RNA (dsRNA) synthesis and injections
Primers specific to different isoforms of Tcdsx containing the T7 promoter sequence at their 5′ ends and cDNA were used to amplify different fragments of Tcdsx. Purified PCR products were used as templates to synthesize dsRNA using MEGAscript T7 kit (Ambion, Austin, TX). A fragment from Escherichia coli malE gene was used to prepare control dsRNA. dsRNA injections were performed into newly eclosed (0 day) pupae. The insects were kept on ice for 8–10 minutes prior to injections. dsRNAs (≈500–600 ng per insect) were injected on the ventral side of the pupae using an aspirator tube assembly (Sigma-Aldrich) fitted with 3.5″ glass capillary tube (Drummond) pulled by a needle puller (Model P-2000, Sutter Instrument Co.). Injected pupae were allowed to recover for 8 hr at room temperature (~22°C) and then were transferred to standard containers containing food. Knockdown efficiencies of gene expression in the RNAi insects were calculated as the ratio of gene expression between 5 day-old (adult) beetles eclosed from pupae injected either with target dsRNA or malE dsRNA.
Quantitative real time PCR
Quantitative PCR was performed using the SYBR Green kit (Roche, USA) according to the manufacturer's instructions. RNA isolation and RT-PCR was done as mentioned above. Three independent biological replicates were included for each treatment. Expression of T. castaneum rp49 gene was used as an endogenous control to normalize the expression data and the gene expression levels were analyzed by 2−ΔΔCt method40.
Imaging and documentation
The ovaries and testes were dissected from the staged insects and stained with acridine orange and the images were taken using Olympus 1×71 Inverted Research Microscope fitted with reflected fluorescence system. Acridine orange was excited using 502 nm laser. “Megna Fire software” (version 1.5) was used to control the microscope, image acquisition and exportation of TIFF files. Figures of all micrographs were assembled using Adobe Photoshop element 9.
References
Bull, J. Evolution of Sex Determining Mechanisms. Menlo Park, Califonia: Benjamin/Cummings Publishing Company (1983).
Zarkower, D. Establishing sexual dimorphism: conservation amidst diversity? Nature Rev Genet. 2, 175–185 (2001).
Verhulst, E. C., van de Zande, L. & Beukeboom, L. W. Insect sex determination: it all evolves around transformer. Current opinion in genet dev. 20, 376–383 (2010).
Salz, H. K. Sex determination in insects: a binary decision based on alternative splicing. Current opinion in genet dev. 21, 395–400 (2011).
Schutt, C. & Nothiger, R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127, 667–677 (2000).
Saccone, G., Pane, A. & Polito, L. C. Sex determination in flies, fruitflies and butterflies. Genetica 116, 15–23 (2002).
Matson, C. K. & Zarkower, D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nature Rev Genet. 13, 163–174 (2012).
Hodgkin, J. The remarkable ubiquity of DM domain factors as regulators of sexual phenotype: ancestry or aptitude? Gene Dev 16, 2322–2326 (2002).
Shukla, J. N. & Nagaraju, J. Doublesex: a conserved downstream gene controlled by diverse upstream regulators. J Genet. 89, 341–356 (2010).
Yi, W. & Zarkower, D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development 126, 873–881 (1999).
Ferguson-Smith, M. The evolution of sex chromosomes and sex determination in vertebrates and the key role of DMRT1. Sexual development: genetics, molecular biology, evolution, endocrinology, embryology and pathology of sex determination and differentiation 1, 2–11 (2007).
Hildreth, P. E. Doublesex, Recessive Gene That Transforms Both Males and Females of Drosophila into Intersexes. Genetics 51, 659–678 (1965).
Burtis, K. C. & Baker, B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997–1010 (1989).
Erdman, S. E. & Burtis, K. C. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J 12, 527–535 (1993).
Erdman, S. E., Chen, H. J. & Burtis, K. C. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144, 1639–1652 (1996).
Burtis, K. C., Coschigano, K. T., Baker, B. S. & Wensink, P. C. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J 10, 2577–2582 (1991).
Salvemini, M. et al. Genomic organization and splicing evolution of the doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol Biol 11, 41 (2011).
Ruiz, M. F. et al. The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. Dev Genes Evol 217, 725–731 (2007).
Lagos, D., Ruiz, M. F., Sanchez, L. & Komitopoulou, K. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348, 111–121 (2005).
Scali, C., Catteruccia, F., Li, Q. & Crisanti, A. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J Exp Biol 208, 3701–3709 (2005).
Concha, C., Li, F. & Scott, M. J. Conservation and sex-specific splicing of the doublesex gene in the economically important pest species Lucilia cuprina. J Genet. 89, 279–285 (2010).
Cho, S., Huang, Z. Y. & Zhang, J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177, 1733–1741 (2007).
Oliveira, D. C. et al. Identification and characterization of the doublesex gene of Nasonia. Insect Mol Biol 18, 315–324 (2009).
Ohbayashi, F., Suzuki, M. G., Mita, K., Okano, K. & Shimada, T. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp biochem physiol B Biochem mol biol 128, 145–158 (2001).
Shukla, J. N. & Nagaraju, J. Two female-specific DSX proteins are encoded by the sex-specific transcripts of dsx and are required for female sexual differentiation in two wild silkmoth species, Antheraea assama and Antheraea mylitta (Lepidoptera, Saturniidae). Insect Biochem Mol Biol 40, 672–682 (2010).
Sugimoto, T. N., Fujii, T., Kayukawa, T., Sakamoto, H. & Ishikawa, Y. Expression of a doublesex homologue is altered in sexual mosaics of Ostrinia scapulalis moths infected with Wolbachia. Insect Biochem Mol Biol 40, 847–854 (2010).
Hediger, M. et al. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol 214, 29–42 (2004).
Shukla, J. N., Jadhav, S. & Nagaraju, J. Novel female-specific splice form of dsx in the silkworm, Bombyx mori. Genetica 139, 23–31 (2011).
Arbeitman, M. N., Fleming, A. A., Siegal, M. L., Null, B. H. & Baker, B. S. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development 131, 2007–2021 (2004).
Chapman, K. B. & Wolfner, M. F. Determination of male-specific gene expression in Drosophila accessory glands. Dev Biol 126, 195–202 (1988).
Wolfner, M. F. Sex-specific gene expression in somatic tissues of Drosophila melanogaster. Trends Genet : TIG 4, 333–337 (1988).
Luo, S. D., Shi, G. W. & Baker, B. S. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138, 2761–2771 (2011).
Coschigano, K. T. & Wensink, P. C. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev 7, 42–54 (1993).
Suzuki, M. G., Funaguma, S., Kanda, T., Tamura, T. & Shimada, T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev 7, 58–68 (2005).
Suzuki, M. G., Funaguma, S., Kanda, T., Tamura, T. & Shimada, T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes and Evol 213, 345–354 (2003).
Kopp, A., Duncan, I., Godt, D. & Carroll, S. B. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559 (2000).
Shirangi, T. R., Dufour, H. D., Williams, T. M. & Carroll, S. B. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol 7, e1000168 (2009).
Wang, W., Kidd, B. J., Carroll, S. B. & Yoder, J. H. Sexually dimorphic regulation of the Wingless morphogen controls sex-specific segment number in Drosophila. Proc Natl Acad Sci USA 108, 11139–11144 (2011).
Hunt, T. et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916, 10.1126/science.1146954 (2007).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Zhang, W. et al. Regulation of sexual dimorphism: mutational and chemogenetic analysis of the doublesex DM domain. Mol Cell Biol 26, 535–547 (2006).
Hedley, M. L. & Maniatis, T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell 65, 579–586 (1991).
Bertossa, R. C., van de Zande, L. & Beukeboom, L. W. The Fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol Biol Evol 26, 1557–1569 (2009).
Posnien, N., Schinko, J., Grossmann, D., Shippy, T. & Konopova, B. RNAi in the red flour beetle (Tribolium). Cold Spring Harb Protoc (2009).
Parthasarathy, R. & Palli, S. R. Molecular analysis of nutritional and hormonal regulation of female reproduction in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol 41, 294–305 (2011).
Parthasarathy, R., Sheng, Z., Sun, Z. & Palli, S. R. Ecdysteroid regulation of ovarian growth and oocyte maturation in the red flour beetle,. Tribolium castaneum. Insect Biochem Mol Biol 40, 429–439 (2010).
Belote, J. M., Handler, A. M., Wolfner, M. F., Livak, K. J. & Baker, B. S. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell 40, 339–348 (1985).
Postlethwait, J. H., Bownes, M. & Jowett, T. Sexual phenotype and vitellogenin synthesis in Drosophila melanogaster. Dev Biol 79, 379–387 (1980).
Parthasarathy, R., Sun, Z., Bai, H. & Palli, S. R. Juvenile hormone regulation of vitellogenin synthesis in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol 40, 405–414 (2010).
Graham, P., Penn, J. K. & Schedl, P. Masters change, slaves remain. BioEssays : news and reviews in molecular, cellular and developmental biology 25, 1–4 (2003).
Chatterjee, S. S., Uppendahl, L. D., Chowdhury, M. A., Ip, P. L. & Siegal, M. L. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development 138, 1099–1109 (2011).
Hoshijima, K., Inoue, K., Higuchi, I., Sakamoto, H. & Shimura, Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252, 833–836 (1991).
Lynch, K. W. & Maniatis, T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev 10, 2089–2101 (1996).
Tian, M. & Maniatis, T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74, 105–114 (1993).
Shukla, J. N. & Palli, S. R. Sex determination in beetles: Production of all male progeny by Parental RNAi knockdown of transformer. Sci. rep. 2, 602 (2012).
Acknowledgements
This work was supported by grants from National Science Foundation (IBN-0421856), National Institutes of Health (GM070559-07) and USDA-NIFA (2011-67013-30143). This is contribution number 12-08-128 from the Kentucky Agricultural Experimental Station.
Author information
Authors and Affiliations
Contributions
Both authors planned and conducted experiments and wrote manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Shukla, J., Palli, S. Doublesex target genes in the red flour beetle, Tribolium castaneum. Sci Rep 2, 948 (2012). https://doi.org/10.1038/srep00948
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00948
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.