Abstract
Imaging modalities including magnetic resonance imaging and X-ray computed tomography are established methods in daily clinical diagnosis of human brain. Clinical equipment does not provide sufficient spatial resolution to obtain morphological information on the cellular level, essential for applying minimally or non-invasive surgical interventions. Therefore, generic data with lateral sub-micrometer resolution have been generated from histological slices post mortem. Sub-cellular spatial resolution, lost in the third dimension as a result of sectioning, is obtained using magnetic resonance microscopy and micro computed tomography. We demonstrate that for human cerebellum grating-based X-ray phase tomography shows complementary contrast to magnetic resonance microscopy and histology. In this study, the contrast-to-noise values of magnetic resonance microscopy and phase tomography were comparable whereas the spatial resolution in phase tomography is an order of magnitude better. The registered data with their complementary information permit the distinct segmentation of tissues within the human cerebellum.
Similar content being viewed by others
Introduction
Minimally or non-invasive surgical interventions on the brain are generally based on less detailed patient-specific imaging (pre- and intra-operative) and high-resolution generic anatomical data. The generic knowledge of brain microstructure currently relies on two-dimensional (2D) evaluation of histological slices, which requires time-consuming sectioning, staining and (manual, expert-based) image analysis. The advantages of histology are sub-micrometer resolution and excellent tunable contrast depending on the staining protocol1,2. However, the technique does not provide isotropic three-dimensional (3D) information of the soft and hard tissues, as is regularly obtained using magnetic resonance imaging (MRI) and X-ray computed tomography (CT). Furthermore, it is hard to imagine image-guided neurosurgery3 without MR guidance. Conventional MRI is a well-established technique for brain imaging and yields superb contrast between white and grey matter but only limited spatial resolution. At present, medical MR systems produce images with typically sub-millimetre voxel lengths. Small animal MR microscopy (µMRI) scanners equipped with stronger gradient systems and operating at higher magnetic field strengths have been used to visualize mouse brains in vivo4,5 and pieces of human brain post-mortem6,7 with voxel sizes of a few tens of micrometers. Conventional CT also provides fully quantitative 3D data ultimately reaching higher spatial resolution than MRI. The CT contrast for brain tissue, however, is weak. Despite low inherent brain tissue contrast, micro-CT (µCT) as a non-destructive technique has already been used to correct deformations induced in data sets obtained by histological sectioning during the preparation of histology8. In order to obtain a better contrast than in these absorption-based studies, synchrotron radiation-based phase-contrast µCT (PC-µCT)9,10 was recently applied to human brain11. X-ray phase-contrast methods are based on the phase shifts of X-ray waves penetrating the specimen which is related to the decrement δ(x,y,z) of the real part of the refractive index distribution. A variety of PC-µCT methods based on crystal interferometry12, propagation-based (or inline) contrast13,14 or analyzer-based imaging (ABI)15,16 are available today. More recently, grating interferometry17,18,19 (also known as Talbot interferometry) was developed, which reaches the especially high contrast required for identifying structures with small differences in electron density as is the case in brain11,20,21.
In the light of these inherent advantages and disadvantages of MR- and CT-based techniques and histology, the aim of the present communication is to elucidate how far the combination of the complementary methods µMRI, grating-based PC-µCT and histology yields additional information on the microanatomy of the human cerebellum.
Results
3D imaging
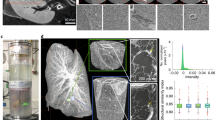
A specimen taken from the human cerebellum (Figure 1) was visualised in 3D using grating-based PC-µCT and µMRI. Figure 2 shows virtual cuts through identical regions of the brain tissue obtained with the two imaging techniques after non-rigid 3D registration22. In the mixed T1/T2*-weighted µMRI images white matter structures appear darker than grey matter structures. Grey matter consists of two components: the stratum granulosum and the stratum moleculare. Because of the weak MR-contrast between these layers, however, it is impossible to segment these structures with an intensity-based algorithm. PC-µCT, on the contrary, provides high contrast between these grey matter layers: with the stratum granulosum appearing as a bright feature and the stratum moleculare as the dark outer structure. The white matter (interior dark region in PC-µCT) exhibits δ-values very similar to the stratum moleculare and can only be segmented because the stratum granulosum is located between the two tissues. Therefore, an intensity-based segmentation is also impossible in this case. This can be deduced from the related histograms of the 3D data in Figure 2. The histogram of the µMRI data (green-coloured triangles in the figure) shows a shoulder (left) associated with white matter and a peak associated with grey matter (right). The histogram of the PC-µCT data (red-coloured squares) contains two peaks and a shoulder. The first peak corresponds to the stratum moleculare, which was in direct contact with formalin during the whole fixation period of the brain. The second peak is a superposition of the stratum moleculare that was in contact with formalin only for restricted periods of time and the white matter. The shoulder on the right originates from the stratum granulosum.
Tissue contrast
The images in Figure 2 acquired with PC-µCT and µMRI show the characteristic morphology of the human cerebellum. The contrast-to-noise ratio c between two anatomical structures can be defined as

where xci are the centre positions of the related peaks in the histograms and FWHMback is the full-width-at-half-maximum of the background signal, here formaldehyde solution and perfluoropolyether, respectively. This definition of the denominator was chosen for practical reasons (see supplementary material). Using Equation 1, six c-values between the four anatomical structures were determined for PC-µCT and µMRI (Figure 3). The highest contrast in the PC-µCT images is the contrast between stratum granulosum and tissue in contact with formalin (E) and has the value of cPC-µCT_max = 6.4 ± 0.2. Using µMRI a maximum value of cµMRI_max = 9.5 ± 0.1 was determined, corresponding to the contrast between white matter and tissue in contact with formalin (C).
Tissue contrast in human cerebellum.
The above data compare contrast values between µMRI and PC-µCT for white matter and stratum granulosum (A), white matter and stratum moleculare (B), white matter and tissue in contact with formalin (C), stratum granulosum and stratum moleculare (D), stratum granulosum and tissue in contact with formalin (E) and between stratum moleculare and tissue with formalin (F).
Joint histogram
The superposition of peaks in the individual histograms prevents the segmentation of the anatomical features. In particular, it is impossible to differentiate between stratum moleculare and stratum granulosum in the µMRI data and between white matter and stratum moleculare in the PC-µCT data. Combining µMRI and PC-µCT using a joint histogram, however, allows the distinct discrimination between the characteristic soft tissues (Figure 4). To do so, the first step was the identification of the 2D maxima in the joint histogram corresponding to the tissues of interest using a multi-Lorentzian fit. Subsequently, the intersections of the Lorentzians were calculated. These intersections characterize the interfaces between the anatomical structures and allow a distinct differentiation between white matter, given in red, stratum granulosum in blue and stratum moleculare in yellow. In addition, the green-coloured peak indicates regions of stratum moleculare, which were in direct contact with formalin during the whole fixation period.
Segmentation by means of a joint histogram.
From the joint histogram of the phase tomography and MR microscopy data it is possible to distinctly segment the stratum granulosum, stratum moleculare, white matter and brain tissue that was in direct contact with formalin for a longer period of time than other parts. The white arrows indicate the positions of the related line plots. The colours of the virtual cuts of PC-µCT and µMRI on the right side correspond to the colours assigned to the four peaks in the joint histogram.
Spatial resolution
In order to evaluate the spatial resolution of µMRI, PC-µCT and histology, firstly line plots through the registered data of the human cerebellum (see Figure 5) were inspected. The spatial resolution of the imaging techniques can be deduced by comparing the roughness of the line plots. Histology has the best spatial resolution followed by PC-µCT then µMRI. To quantify this observation, the spatial resolution λSR was calculated by Fourier analysis11,23. The radial spectral power (rSP) of a structure of interest in a 100 × 100 pixel region of a selected slice (rSPstruc) was calculated and divided by the rSP of a background region of same size (rSPback) (i.e. of formalin for PC-µCT, perfluoropolyether for µMRI and glass without brain tissue for histology). This ratio was plotted against the spatial frequency. The peak-to-peak distance of the first frequency larger than twice the mean value of the baseline (noise) was divided by two to determine the spatial resolution24. The λSR-values corresponded to (6 ± 1) µm for histology, to (20 ± 5) µm for PC-µCT and to (210 ± 40) µm for µMRI.
Multimodality
The morphology of the cerebellum can be properly visualized combining µMRI, PC-µCT and histology. Figure 6 illustrates one possible way of combining the 2D images of the three techniques used in this study. Each technique is represented by one of the colour channels red/green/blue (RGB) (see Figure 6, left image). Such a colour representation directly illustrates the contributions of the individual techniques. For example the yellow colour, which indicates the stratum granulosum, is mainly the superposition of contrast from the PC-µCT and µMRI, whereas the purple colour (white matter) primarily arises from the PC-µCT and the histology signals.
Multimodal imaging of the cerebellum.
The fusion of slices from PC-µCT, µMRI and histology using the RGB channels directly visualizes the contributions from the three imaging techniques. The combination of the three techniques permits the distinction of otherwise indistinguishable anatomical features, such as the separation between white matter (violet), stratum granulosum (yellow), stratum moleculare (green) and blood vessels of different sizes (purple and white arrows). The enlarged regions (with different magnification) show the light blue coloured small capillaries.
Discussion
Grating interferometry is based on the detection of the deflection angles of the X-rays passing through the specimen. After reconstruction, a 3D distribution of the decrement δ of the real part of the refractive index of the specimen can be obtained, which is directly related to the electron density distribution

with the classical electron radius re and the X-ray wavelength λ25.
In MRI, contrast between different tissues is determined by their relative spin densities ρs, their characteristic relaxation properties at the purported magnetic field strength, i.e. their distinguished T1 (longitudinal) and T2 resp. T2* (transversal) relaxation times and the imaging sequence and parameters used (in particular the echo time TE, the repetition time TR and the flip angle α). In a very general manner, the measured amplitude of the signal for a gradient echo sequence can be described as

where

takes into account the refocusing of magnetization components that were dephased under the readout gradient of the imaging sequence, while dephasing of magnetization resulting from static field inhomogeneities such as susceptibility effects contribute to the image contrast. In order to optimize tissue contrast, dedicated imaging sequence parameters were chosen based on literature values for grey and white matter at magnetic fields of 9.4 T26.
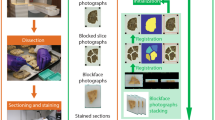
Before the acquired data can be compared and combined, they must be registered with each other in 3D space. It was shown that the 3D registration using the maximization of the mutual information22 gives precise results for the registration of tomography data from different imaging modalities27. A rigid registration including three degrees of translation and three degrees of rotation seems to be sufficient for the 3D data set acquired by PC-µCT and µMRI. Although the brain tissue undergoes shrinkage during formalin fixation, further deformations over long-time periods are minimal. Nevertheless, an affine registration with an additional three scaling factors compensates for the error in individual pixel sizes. In our case a non-rigid registration was required to correlate the corresponding voxels between the imaging modalities. This approach accounts for possible deformations within the brain tissue induced during the transfer of the specimen from the cylindrical polyethylene container (filled with formaldehyde) used in PC-µCT measurements into the container (with perfluoropolyether) for µMRI. Because of the deformations induced during sectioning and staining, the histological slice was also non-rigidly registered with the tomography data.
The virtual cuts through the µMRI and PC-µCT data sets in Figure 2 elucidate the pros and cons of each method. It is well known that MRI provides superb contrast between white and grey matter. White matter (dark region) can easily be distinguished from the grey matter (bright region) in our mixed T1/T2*-weighted µMRI images. On the contrary, differentiation between layers of grey matter tissues, in particular between stratum granulosum and stratum moleculare, is hardly possible. Here, PC-µCT provides a clear distinction between these anatomical structures. White matter (inner dark region), however, has a comparable electron density to stratum moleculare (outer dark region). These observations are quantified using Equation 1 and summarized in Figure 3. With the definition of the contrast-to-noise ratio c presented in this manuscript, a value c > 1 is required to distinguish anatomical structures using intensity-based algorithms. Therefore, one can conclude that the PC-µCT contrast is sufficiently high to distinguish between the four tissues whereas µMRI contrast is restricted to white and grey matter. Additionally, the PC-µCT pixel size is a factor of nine smaller than µMRI, although the maximal c-value observed in the PC-µCT results is slightly lower than the maximal µMRI contrast-to-noise ratio. Binning the tomography data reduces the spatial resolution but improves the contrast28. Therefore, we can state that the contrast-to-noise between the two methods yields comparable values.
In addition to the contrast-to-noise ratio c, we also consider the spatial resolution λSR to assess image quality. Following the work of Thurner et al.28, we combine these two values in the quality factor q

Using the maximal contrast values of PC-µCT and µMRI and the corresponding spatial resolution values, one obtains quality factors of (0.32 ± 0.09) µm−1 for PC-µCT and (0.05 ± 0.01) µm−1 for µMRI. Based on this definition, PC-µCT yields six times better images than µMRI. More importantly, however, the complementary results of the two data sets can be derived using a joint histogram as presented in Figure 4. Three anatomical structures can be segmented, i.e. white matter (red), stratum granulosum (blue) and stratum moleculare (yellow). Furthermore, due to its high sensitivity, PC-µCT allows the detection of differences within the tissue induced by different periods of formalin fixation. The green regions in Figure 4 correspond to domains of stratum moleculare, which were in direct contact with formalin for the whole fixation period of 120 days and not only for 20 days.
In summary, we demonstrate the advantages and disadvantages of PC-µCT, µMRI and histology and present a method to combine data from these complementary techniques in order to segment the anatomical layers of the human cerebellum post mortem. The clear segmentation of the three layers allows us to quantify the ratios between these volumes. Given that cell death in the cerebellum is a common cause of neurological disease, e.g. in cerebellar ataxias29 and that abnormalities in the cerebellum have more recently also been implicated in autism and neuropsychiatric disorders30,31 methods for a reliable quantitative assessment of cerebellar morphology and pathology are certainly needed. Unfortunately, the currently available stereological methods for the cerebellum are very time consuming and often lack precision31,32. The presented method has the potential to significantly reduce the effort and improve the quality of quantitative stereological assessments of the cerebellum.
Methods
Specimen preparation
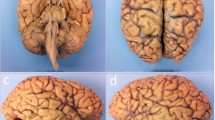
The brain from the donated body of a 68-year-old male was extracted at the Institute of Anatomy (University of Basel, Switzerland) within 48 h after death. The whole brain was transferred to 10% formalin solution for fixation. All procedures were conducted in accordance with the Declaration of Helsinki and according to the ethical guidelines of the Canton of Basel. The small cerebellum block (6 × 6 × 11 mm3) was extracted at the University Hospital Zurich, Switzerland, after approximately three months of fixation (Figure 1). During the measurements the specimen was placed in a 0.5 mL Eppendorf container filled with 4% formalin solution for PC-µCT and with perfluoropolyether (Fomblin® Y-LC 80, Solvay Solexis, Bollate, Italy) for µMRI in order to reduce the background signal and susceptibility artefacts at the tissue-liquid interface33.
Grating-based X-ray phase tomography
A detailed description of grating interferometry can be found in the literature11,19,34,35. The experiment was performed at the beamline ID19 (ESRF, Grenoble, France) at the interferometer installed there36 with X rays from the U32 undulator with a gap set to 15.15 mm. Using a double-crystal Si(111)-monochromator in Bragg geometry a photon energy of 23 keV was selected. The imaging of the entire specimen required two scans to obtain a field of view 11 mm high and 6 mm wide. The beam-splitter grating g1 (made at the Paul Scherrer Institut, Villigen, Switzerland) with a periodicity of p1 = 4.785 μm had a Si structure height of 29 μm, which corresponds, within a few percent, to a phase shift of π for 23-keV X rays. The analyzer grating g2 (made at the Karlsruhe Institute of Technology, Eggenstein-Leopoldshafen, Germany) had lines of gold with a periodicity of p2 = 2.400 μm and 50 µm structure height. This corresponds to an intensity transmission of only about 6 × 10−3 through the teeth of the analyzer grating comb, i.e., a contrast of 100%. With a distance between source and interferometer of 150 m and a distance between the gratings of d = 479.4 mm (corresponding to the 9th Talbot order), the ratio of the grating periods p2/p1 was matched to the beam divergence35. The sample container was immersed in a water tank with parallel polymethylmethacrylate plates in order to minimize artefacts due to X-ray phase curvature induced by a conical container-air interface. The specimen was located 10 cm upstream of the beam-splitter grating. The detector was a lens-coupled scintillator and charge-coupled device (CCD) system using a FreLoN 2K (Fast-Readout, Low-Noise, ESRF Grenoble, France) CCD with 2048 × 2048 pixels and an effective pixel size of 5.1 µm, placed approximately 3 cm downstream of the analyzer grating. Projection radiographs were taken in 1501 steps over a range of 360°. At each projection angle, four phase-stepping images with an exposure time of 1 s each were taken over one period of the interference pattern.
Magnetic resonance microscopy
The µMRI measurements were carried out on a 9.4 T, 30 cm horizontal small animal MR unit (Bruker BioSpec, Bruker BioSpin MRI, Ettlingen, Germany) equipped with a transceiver cryogenic quadrature radio frequency (RF) surface coil (CryoProbeTM) designed for MRI of the mouse brain at increased sensitivity. High-resolution structural images were acquired with a T2*-weighted 3D FLASH sequence with isotropic voxel size of 45 µm (field of view (FOV) 1.35 × 0.90 × 1.35 mm3, acquisition matrix (MTX) 300 × 200 × 300, echo time (TE) 12 ms repetition time (TR) 400 ms, flip angle (α) 15°, number of averages (NA) 1 at a total acquisition time of 400 minutes.
Histology
Sections of 50 µm thickness of the cerebellum block were prepared using a microtome with vibrating blade HM 650 V (Microm International GmbH, Walldorf, Germany). For the staining procedure, the sections were mounted on gelatinized slides and stained for Nissl with cresyl violet1. Microphotographs of selected slices were taken using a Leica MZ16 microscope and DFC420-C digital camera.
Data processing
The reconstruction of the phase-contrast dataset was carried out using a modified filter kernel (Hilbert transform) in combination with a standard back-projection algorithm34,37,38. Because the signal intensity in the µMRI images varies with the coil sensitivity profile, further processing was performed. For the correction of the inhomogeneities a modified fuzzy C-means algorithm was applied39. For a reasonable analysis of the joint histogram, the 3D datasets were non-rigidly registered using the classical maximization of mutual information (MI) principle40,41. The chosen registration algorithm is based on the adaptive hierarchical image subdivision strategy, which decomposes the non-rigid matching problem into numerous local affine registrations of image segments of decreasing size22. The local registrations were again performed using the classical maximization of MI principle. The local registration parameters were identified using the Powell multi-dimensional search algorithm42 such that the MI between the reference and the floating sub-images was maximized. The hierarchical image splitting strategy43 recently underwent several improvements and was extended to 3D22,44. For region selection in the joint histogram, the line plots were approximated with a multi-Lorentzians fit using the Levenberg–Marquardt algorithm in OriginPro 7.5 (OriginLab Corporation, Northampton, USA).
References
Morel, A. Stereotactic Atlas of the Human Thalamus and Basal Ganglia. (Informa Healthcare New York, 2007).
Schaltenbrand, G. & Wahren, W. Atlas for Stereotaxy of the Human Brain. (Thieme, Stuttgart, 1977.
Jolesz, F. A., Nabavi, A. & Kikinis, R. Integration of interventional MRI with computer-assisted surgery. J. Magn. Reson. Imaging 13, 69–77 (2001).
Benveniste, H. & Blackband, S. MR microscopy and high resolution small animal MRI: Applications in neuroscience research. Prog. Neurobiol. 67, 393–420 (2002).
Ahrens, E. T. et al. MR microscopy of transgenic mice that spontaneously acquire experimental allergic encephalomyelitis. Magn. Reson. Med. 40, 119–132 (1998).
Fatterpekar, G. M. et al. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. Am. J. Neuroradiol. 23, 1313–1321 (2002).
Fatterpekar, G. M. et al. MR microscopy of normal human brain. Magn. Reson. Imaging Clin. N. Am. 11, 641–653 (2003).
Germann, M. et al. Strain fields in histological slices of brain tissue determined by synchrotron radiation-based micro computed tomography. J. Neurosci. Methods 170 (1), 149–155 (2008).
Fitzgerald, R. Phase-sensitive X-ray imaging. Phys. Today 53 (7), 23–26 (2000).
Momose, A., Fujii, A., Kadowaki, H. & Jinnai, H. Three-dimensional observation of polymer blend by X-ray phase tomography. Macromolecules 38 (16), 7197–7200 (2005).
Schulz, G. et al. High-resolution tomographic imaging of a human cerebellum: Comparison of absorption and grating-based phase contrast. J. R. Soc. Interface 7, 1665–1676 (2010).
Bonse, U. & Hart, M. An X-ray interferometer. Appl. Phys. Lett. 6 (8), 155–156 (1965).
Snigirev, A. A., Snigireva, I., Kohn, V., Kuznetsov, S. & Schelokov, I. On the possibilities of x-ray phase contrast microimaging by coherent high-energy synchrotron radiation. Rev. Sci. Instrum. 66 (12), 5486–5492 (1995).
Cloetens, P., Barrett, R., Baruchel, J., Guigay, J.-P. & Schlenker, M. Phase objects in synchrotron radiation hard x-ray imaging. J. Phys. D: Appl. Phys. 29, 133–146 (1996).
Förster, E., Goetz, K. & Zaumseil, P. Double crystal diffractometry for the characterization of targets for laser fusion experiments. Krist. Tech. 15 (8), 937–945 (1980).
Davis, T. J., Gao, D., Gureyev, T. E., Stevenson, A. W. & Wilkins, S. W. Phase-contrast imaging of weakly absorbing materials using hard X-rays. Nature 373, 595–598 (1995).
David, C., Nöhammer, B., Solak, H. H. & Ziegler, E. Differential X-ray phase contrast imaging using a shearing interferometer. Appl. Phys. Lett. 81, 3287–3289 (2002).
Momose, A. et al. Demonstration of X-ray Talbot interferometry. Jpn. J. Appl. Phys., Part 2 42 (7B), 866–868 (2003).
Weitkamp, T. et al. X-ray phase imaging with a grating interferometer. Opt. Express 13, 6296–6304 (2005).
Pfeiffer, F. et al. High-sensitivity phase-contrast tomography of rat brain in phosphate buffered saline. J. Phys. Conf. Ser. 186, 012046 (2009).
Pfeiffer, F. et al. High-resolution brain tumor visualization using three-dimensional X-ray phase contrast tomography. Phys. Med. Biol. 52 (23), 6923–6930 (2007).
Andronache, A., von Siebenthal, M., Székely, G. & Cattin, P. Non-rigid registration of multi-modal images using both mutual information and cross-correlation. Med. Image Anal. 12 (1), 3–15 (2008).
Modregger, P., Lübbert, D., Schäfer, P. & Köhler, R. Spatial resolution in Bragg-magnified X-ray images as determined by Fourier analysis. Phys. Status Solidi A 204, 2746–2752 (2007).
Wang, L., Ho, P. P., Liu, C., Zhang, G. & Alfano, R. R. Ballistic 2-D imaging through scattering walls using an ultrafast optical Kerr gate. Science 253, 769–771 (1991).
Als-Nielsen, J. & McMorrow, D. Elements of Modern X-ray Physics. (John Wiley & Sons, Chichester, 2011).
Kuo, Y.-T., Herlihy, A. H., So, P.-W., Bhakoo, K. K. & Bell, J. D. In vivo measurements of T1 relaxation times in mouse brain associated with different modes of systemic administration of manganese chloride. J. Magn. Reson. Imaging 21, 234–239 (2005).
Müller, B. et al. Three-dimensional registration of tomography data for quantification in biomaterials science. Int. J. Mater. Res. 103, 242–249 (2012).
Thurner, P., Beckmann, F. & Müller, B. An optimization procedure for spatial and density resolution in hard X-ray micro-computed tomography. Nucl. Instrum. Methods Phys. Res. B 225, 599–603 (2004).
van Gaalen, J. & van de Warrenburg B.P.C. A practical approach to late-onset cerebellar ataxia: putting the disorder with lack of order into order. Pract. Neurol. 12, 14–24 (2012).
Villanueva, R. The cerebellum and neuropsychiatric disorders. Psychiatry Res. in press, published online before print 22 March (2012).
Andersen, K., Andersen, B. B. & Pakkenberg, B. Stereological quantification of the cerebellum in patients with Alzheimer's disease. Neurobiol. Aging 33, 197.e111–197.e120 (2010).
Agashiwala, R. M., Louis, E. D., Hof, P. R. & Perl, D. P. A novel approach to non-biased systematic random sampling: A stereologic estimate of Purkinje cells in the human cerebellum. Brain Res. 1236, 73–78 (2008).
Smith, B. R., Huff, D. S. & Johnson, G. A. Magnetic resonance imaging of embryos: An internet resource for the study of embryonic development. Comp. Med. Imag. Graph. 23, 33–40 (1999).
Pfeiffer, F., Kottler, C., Bunk, O. & David, C. Hard X-ray phase tomography with low-brilliance sources. Phys. Rev. Lett. 98, 108105 (2007).
Weitkamp, T., David, C., Kottler, C., Bunk, O. & Pfeiffer, F. Tomography with grating interferometers at low-brilliance sources. Proc. SPIE 6318, 63180S (2006).
Weitkamp, T. et al. Recent developments in X-ray Talbot interferometry at ESRF-ID19. Proc. SPIE 7804, 780406 (2010).
Faris, G. W. & Byer, R. L. Three-dimensional beam-deflection optical tomography of a supersonic jet. Appl. Opt. 27, 5202–5212 (1988).
Pfeiffer, F., Bunk, O., Kottler, C. & David, C. Tomographic reconstruction of three-dimensional objects from hard X-ray differential phase contrast projection images. Nucl. Instrum. Methods Phys. Res. Sect. A 580, 925–928 (2007).
Ahmed, M. N., Yamany, S. M., Mohamed, N., Farag, A. A. & Moriarty, T. A modified fuzzy C-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans. Med. Imaging 21, 193–199 (2002).
Maes, F., Collignon, A., Vandermeulen, D., Marchal, G. & Suetens, P. Multi-modality image registration by maximization of mutual information. Mathematical Methods in Biomedical Image Analysis. IEEE, 14–22 (1996).
Viola, P. & Wells, W. M. Alignment by maximization of mutual information. Proceedings of the fifth international conference on computer vision, 16–23 (1995).
Press, W. H., Flannery, B. P., Teukolsky, S. A. & Vetterling, W. T. Numerical Recipes in C - The Art of Scientific Computing. (Cambridge University Press, 1988).
Likar, B. & Pernus, F. A hierarchical approach to elastic registration based on mutual information. Image Vis. Comput. 19 (1–2), 33–44 (2001).
Schulz, G. et al. Three-dimensional strain fields in human brain resulting from formalin fixation. J. Neurosci. Methods 202, 17–27 (2011).
Acknowledgements
The authors gratefully acknowledge M. Müller-Gerbl and P. Zimmermann for the organization and carrying out the extraction of the human brain, J. Kapfhammer for anatomical and medical information, A. Morel for the brain specimen blocking, M. Imholz for the histology of the specimen, S. Rutishauser for the preparation of the beam-splitter grating, M. Rudin for giving us the opportunity to perform the magnetic resonance microscopy experiments and A. Andronache for making the non-rigid registration tool available. The authors thank M. N. Holme for improving the readability and for correcting the language. T. W. acknowledges support from the French research networks (réseau thématique de recherche avancée, RTRA) “Digiteo” and “Triangle de la Physique” (grants 2009-034T and 2009-79D). F. P. acknowledges financial support through the DFG Cluster of Excellence Munich-Center for Advanced Photonics and the European Research Council (FP7, Starting Grant No. 240142). The project was partially funded by the Swiss National Science Foundation (CR23I2_125 406) and was supported by the ESRF (proposal MD-407) through beam time allocation. This work was partly conducted with the support of the Karlsruhe Nano Micro Facility (KNMF), a Helmholtz Research Infrastructure at Karlsruhe Institute of Technology (KIT).
Author information
Authors and Affiliations
Contributions
G.S. prepared the specimen for the measurements; F.P., T.W. and C.D. developed the phase-contrast set up; G.S., I.Z., T.W. and B.M. performed phase-contrast tomography experiments; C.W. performed magnetic resonance microscopy experiments; G.S. and B.M. analyzed the data and prepared the figures; G.S. and B.M. wrote the initial manuscript; all authors discussed the results and contributed to the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Schulz, G., Waschkies, C., Pfeiffer, F. et al. Multimodal imaging of human cerebellum - merging X-ray phase microtomography, magnetic resonance microscopy and histology. Sci Rep 2, 826 (2012). https://doi.org/10.1038/srep00826
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00826
This article is cited by
-
Quantitative X-ray phase contrast computed tomography with grating interferometry
European Journal of Nuclear Medicine and Molecular Imaging (2021)
-
Optimising complementary soft tissue synchrotron X-ray microtomography for reversibly-stained central nervous system samples
Scientific Reports (2018)
-
A simple setup for episcopic microtomy and a digital image processing workflow to acquire high-quality volume data and 3D surface models of small vertebrates
Zoomorphology (2018)
-
X-ray-Based 3D Virtual Histology—Adding the Next Dimension to Histological Analysis
Molecular Imaging and Biology (2018)
-
Experimental Realisation of High-sensitivity Laboratory X-ray Grating-based Phase-contrast Computed Tomography
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.