Abstract
Automated image analysis of cells and tissues has been an active research field in medical informatics for decades but has recently attracted increased attention due to developments in computer and microscopy hardware and the awareness that scientific and diagnostic pathology require novel approaches to perform objective quantitative analyses of cellular and tissue specimens. Model-based approaches use a priori information on cell shape features to obtain the segmentation, which may introduce a bias favouring the detection of cell nuclei only with certain properties. In this study we present a novel contour-based “minimum-model” cell detection and segmentation approach that uses minimal a priori information and detects contours independent of their shape. This approach avoids a segmentation bias with respect to shape features and allows for an accurate segmentation (precision = 0.908; recall = 0.859; validation based on ∼8000 manually-labeled cells) of a broad spectrum of normal and disease-related morphological features without the requirement of prior training.
Similar content being viewed by others
Introduction
The earliest attempts to use computers for the automated detection and analysis of cells in cyto-/histological specimens date back a few decades1,2. Since then the topic has established itself as an important area of research in biomedical informatics3,4,5,6,7,8. However, as the more recent literature shows the search for a robust, practically usable cell/nucleus segmentation module is still ongoing: Various different approaches have been proposed ranging from relatively simple thresholding techniques9,10 to more sophisticated methods, such as, for instance, adaptive attention windows defined by the maximum cell size11 or gradient flow tracking12. A “2D world decomposition” model combined with a priori texture and color information is used for the segmentation of specific cells (leukocytes) in microscopic images of bone marrow13. In14 stepwise merging of rules based on mean-shift clustering and boundary removal rules with a gradient vector flow snake is used for the segmentation of blood cells. Another very recent study15 uses a fuzzy C-means active contour algorithm that combines multispectral edge and region information through a vector multiphase level set framework. Further approaches including the separation of cell clusters can be found in16,17,18,19,20,21. This selection of publications exemplifies the methodological diversity employed to tackle this complex problem. However, despite these long-standing efforts in computer science it is only very recently that automated cell segmentation has also received increased attention on the application side in clinical pathology. The reasons for this are on one hand technology-based: Only in the last years computers, microscope cameras and whole slide scanners have become widely available and are able to generate, process and analyze high-quality digitized histological data sets. The other reasons are biomedical: While qualitative or at most semi-quantitative “visual” estimation of morphological features (for instance nuclear shape in cancer cells as a proxy of the grade of malignancy) or the expression of immunohistological markers are still considered the standard approach in diagnostic pathology, the requirements for cyto- and histological analyses are currently changing. More and more prognostic and predictive markers, i.e. markers that enable pathologists to assess the aggressiveness of a disease or its response to therapy, respectively, are being discovered with increasing requirements for standardized quantitative assessment22,23,24,25. Here, automated image analysis tools will not replace but assist pathologists to increase diagnostic precision and inter-observer reliability. Examples of potential applications of “computer-assisted diagnostics” include, for instance, the scoring of growth factor receptor expression levels in gastric and breast cancer to decide if a patient will benefit from antibody-based inhibitory therapy26,27, the quantification of nuclear shape features in cancer cells (“grade”)28or the quantification of the tumor proliferation index as measured by Ki-67 immunohistochemistry29. Independent of the particular task to be performed all approaches share one algorithmic module pivotal for the accurate performance of any cellular segmentation and analysis approach: the robust detection (including localization and segmentation) of cell nuclei. To achieve this goal existing approaches often use a priori information about cellular/nuclear features13,14, which are used to build a generic cell model3,5,7,8. Such approaches perform well if cell populations are relatively homogeneous in terms of shape or size. However, especially in the pathologically most relevant cases, i. e. the segmentation of cancer cell nuclei in cytology or histology samples, this approach is problematic in two ways, because cancer cells have highly heterogeneous shape and size features and moreover are accompanied by stromal and immune cells of various shapes and sizes: On one hand this makes it difficult to define a generic contour model representative of a realistic distribution of cell nucleus features. Second and this aspect is even more relevant, using a priori information and generating a model of contour features has a high risk of introducing a segmentation bias: Nuclei with extreme features would have a higher probability of not being covered by the model parameters and might thus be overlooked in comparison with more “regular” cell nuclei. Because cells with extreme shape deviations with respect to a “model cell” are very often the most malignant ones, such a bias might lead to significant diagnostic errors in all of the above mentioned applications and is therefore not acceptable. The only way around this issue would be to use very loose model parameters, which would, however, result in an unacceptably high number of false positives. In the study presented here, we describe a contour-based nucleus detection algorithm, which uses minimal a priori information and finds contours independent of their shape by combining a global contour search with local gradient information. This approach avoids a segmentation bias with respect to shape features and is capable of segmenting cell nuclei of various normal and cancerous cell types from breast, liver and bone marrow as well as kidney tissue and intestinal mucosa with a single set of parameters and no prior training.
Results
The cell nucleus segmentation approach we present in this study consist of six major modules: First, all possible closed contours present in the grayscale-transformed image are computed regardless of size, shape or whether they belong to hills or valleys in the intensity landscape. This step yields multiple, often overlapping contours, which are evaluated in a second module based on gradient features of the input image. We introduce a “contour value” as a measure to rank and select those contours that best represent the image objects. The contour value is a combination of the mean contour gradient that measures the relative “importance” of objects (in a group of overlapping objects) and the “gradient fit” that defines which of several alternative contours best represents a certain object. A non-overlapping segmentation is generated in the third step: The enclosed areas of the ranked contours are labeled in a two dimensional map. Subsequently, this segmentation is improved using a novel contour optimization method and an optional cluster separation step. Finally, cell nuclei are detected by assessing the (nucleus-specific) Hematoxylin within each contour area (for details see Methods section). To quantitatively evaluate the performance of the cell/nucleus detection we prepared a gold-standard data set using randomly-selected images containing 7931 cells manually labeled by a pathologist. The selected tissues comprise various breast cancer samples representing a broad morphological variety including normal tissue components, bone marrow with normal and pathologically altered cells, normal liver tissue as well as kidney tissue and intestinal mucosa acquired during routine diagnostics and processed with standard routine laboratory protocols at our institute (see Methods for details). After the manual labeling, the original image data was segmented using the proposed method and the positional information available in the gold-standard data was compared to the automated segmentation result by automatically determining true positives, false negatives and false positives. This analysis yielded an overall precision of 0.908, a recall of 0.859 and a conglomerate score of 0.953. These data feature a broad range of cell/nucleus morphologies ranging from normal gastric mucosa (regular round/oval shaped nuclei) to bone marrow with primary myelofibrosis (highly irregular neoplastic megacaryocytes, Fig. 1d and Fig. 33 in Supplementary Notes S1 online). The analysis of these images shows similar segmentation accuracies irrespective of the different morphologies (Myelofibrosis: precision = 0.88, recall = 0.91; normal mucosa: precision = 0.90, recall = 0.89), which confirms the absence of a segmentation bias in our method. A comparison to Al-Kofahis method30,31 was performed using first the default (fully automatic) mode (which was recommended by the authors upon request) and second, using an optimized parameter configuration file provided by the authors after submitting test data to them (Table 1). When using the automatic mode Al-Kofahis method required less processing time but yielded a by 0.197 lower precision (p = 5.97e-10) and by 0.049 higher recall (p = 2.03e−7). With optimized parameters the precision remained 0.085 less precise than our approach (p = 5.60e−7) with a constant recall (0.908, p = 2.24e−7) and requiring 2.6 times more mean processing time compared to our approach. The conglomerate score of Al-Kofahis method was only slightly better: 0.953 in our approach compared to 0.964 (p = 3.75e−3) in the automatic mode and 0.966 (p = 1.16e−3) with optimized parameters. All images (original and labeled) and the individual segmentation accuracies can be found side-by-side in Supplementary Note S1. Moreover, Supplementary Data S2 contains the original image data and the xml-coded positional information from the manual cell labeling.
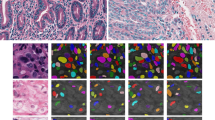
H&E stained tissue samples: (a) breast cancer; (b) liver; (c) gastric mucosa; (d) bone marrow with primary myelofibrosis with highly pleomorphic megakaryocytes (without cell cluster separation); (e) connective tissue; (f) kidney tissue; (e, f) examples of the method validation (dots represent manually-assigned labels, green: labels classified as true positive, red: false negatives, yellow: false positives).
Discussion
Research in automated cell nucleus segmentation has recently regained attention because of an increased interest by the pathology community in standardized and quantitative evaluation of cyto- and histological specimens. This is on one side due to an increasing number of immunohistochemical diagnostic markers that need to be quantified to allow for a valid prognostic and/or predictive assessment. Moreover, the use of nuclear shape features for prognostic purposes in cancer diagnostics has recently received criticism due to a large inter-observer variability, especially in the context of novel genomic test (such as, for instance, OncotypeDX or MammaPrint) that compete with classical histopathology in the growing field of individualized predictive pathology. In this study, a contour-based segmentation method is described that is closely simulating the way the human eye detects cells, i. e. mainly based on the extent the objects are silhoutted agains the background. Also, contours are detected independent of their shape. Consequently, very pleomorphic cell nuclei are found with the same accuracy as very “normal”, i. e. round cell nuclei and even contours with a contour-gradient of only one intensity value (difference between the objects contour and its surrounding) may be detected if these are the contours with the locally strongest contour-gradient. The method uses minimal a priori information about cell nucleus staining and size. Compared to the approach by Al-Kofahi et al.30,31, that uses 9 parameters, our method requires only 5 parameters that may be set within relatively loose bounds. Performing segmentation with few model assumptions on cell shape features can help avoid the introduction of a detection bias which would favour normal cells without nuclear atypia over pleomorphic malignant cells and therefore lead to significant diagnostic errors especially in the case of cancer tissues that display large morphological varieties. Our approach is capable of segmenting cell nuclei of various normal and cancerous cell types from breast, liver and bone marrow and other tissues with a single set of parameters and no prior training. Moreover, the described features render the method robust against variations in staining protocols within reasonable limits, because no absolute intensity information is used initially for contour detection. The method is also relatively robust against image blur (see Supplementary Note S3 online). Due to the general (minimum) model formulation our method is also capable of segmenting fluorescence microscopy data (e. g. confocal laser scanning microscopy images) or even electron microscopy images. It may therefore in the future also be useful for the registration of the different imaging modalities in the field of correlative light electron microspy (CLEM)32,33 (see Supplementary Note S4 online). Our method is therefore highly versatile and suitable to serve as a basis in form of a segmentation module for applications, for instance, designed to assess cancer malignancy (pathological grading) for which the variability of nucleus shapes is a central feature, or the immunohistochemical quantification of the proliferation index by immunostaining of the Ki-67 antigen, for which it is critical to detect all variants of cancer cells. In case of strong unspecific background Hematoxylin staining, false positive cell nuclei may occur, which, however, is rarely the case in the data we analysed. As our visual tests and quantitative validation with more than 7900 manually labeled cells show the proposed method performs well for a large variety of morphological cancer variants and normal tissue exhibiting a representative variation of nucleus shape features. To conclude, we propose a novel cell nucleus segmentation approach that minimizes the use of a priori knowledge and may therefore serve as the basis for segmentation modules in cancer nucleus feature quantification and classification tasks.
Methods
The cell nucleus segmentation approach presented here consists of six internal processing steps:
-
a
Detection of all possible closed contours
-
b
Contour evaluation
-
c
Generation of a non-overlapping segmentation
-
d
Contour optimization
-
e
Concave object separation
-
f
Classification into cell nuclei and other objects
Detection of all possible contours (a)
To find the best fitting contour for each nucleus within an image, initially, our method detects all possible closed contours irrespective of whether they actually belong to a nucleus or not. We call this the primary segmentation.
This contour detection is based on a conventional contour tracing approach for binary images34, which we extended for the use with grayscale images. The grayscale image is generated by computing the mean of the red, green and blue channel of the corresponding pixel in the original color image. While finding the contour start pixels (seeds) needed to initiate the contour tracing is straightforward for binary images, grayscale images require a different strategy: in our approach each image row is regarded as a one dimensional image function I(x) and the contour start pixels are defined as the positions at which the gradient between each pair of neighboring local minima/maxima or maxima/minima is maximal (Fig. 2). All pixels with grayscale values between the intensity at the contour start pixel and at the corresponding local minimum or the corresponding local maximum, respectively, are initially interpreted as object-pixels (Fig. 3). As a consequence each contour tracing is performed within a locally-adapted (“individual”) intensity range and objects may represent either “hills” or “valleys” in the intensity landscape. The detection of the contour start pixels and the determination of the corresponding intensity range is performed by scanning the image row-wise from left to right and storing all local minima and maxima with the corresponding maximum gradients in between (visual tests showed that the overall result is invariant with respect to the scan direction). Subsequently, the contour tracing follows the (potential) object contours clockwise using an 8-connected neighborhood (according to34). A contour is considered valid if and only if it reaches back to its start pixel. Otherwise, the contour tracing is terminated if a maximum contour length (225 pixels in our examples) is exceeded. Pseudocode for the contour start pixel and extremal value detection and an english description of the contour tracing algorithm can be found in the Supplementary Methods S5 and S6 online.
Contour evaluation (b)
The contour detection approach (primary segmentation) results in multiple, often overlapping contours many of which do not represent actual objects at all. Thus, this primary segmentation is not composed of disjunct image segments and therefore heavily over-defined. To obtain a non-overlapping segmentation it has to be determined, which objects are more prominent than others within the same local area: An object is considered more important if it has a higher mean gradient along the objects contour (Equation 3), which largely corresponds to the (trivial) concept that an image region is more likely to correspond to an actual object if it is silhouetted sharply against the background (or other objects). Additionally, multiple contours from the primary segmentation may describe the same object and hence the decision has to be made, which of several alternative contours best represents a certain object: Contours are visually regarded as fitting best if they show a high concordance between contour pixels and the maximum local gradient. This “gradient fit” is defined as the relative number of contour pixels that represent a local maximum in a 3×3 neighborhood in the gradient image (Equation 2) and is computed by using the Sobel operator S with its 3×3 convolution kernels Gx and Gy (Equation 1) and the 2-D image function I. The mean gradient and the “gradient fit” are combined in the contour value (Equation 4). In equation (2) and (3) let Ci be the ith contour and pij its jth contour pixel.




Generation of a non-overlapping segmentation (c)
To generate a non-overlapping segmentation, initially, a two-dimensional map of the same size as the corresponding image is used to label the area within each contour with a unique identifier. To obtain the locally most prominent contours (as defined by the contour value defined in Eq. 4) the labeling process is performed in sorted order, beginning with the topmost valued contour and blocking overwriting of already assigned labels.
Contour Optimization (d)
Due to the consecutive labeling process objects may have thin filaments on the object border (Fig. 4b). These filaments do not belong (are not compact) to the actual object. To remove these filaments a contour optimization processing step is applied (Fig. 4c): The distance transformation is computed based on the chamfering method introduced by35 using the Manhattan metric. A distance value d is defined for testing the compactness of object pixels (3 in our examples). A Pixel pi (with di the distance value of pi and di < d) is considered “compact” if it is connected to a pixel pj (with dj = d) over d − di edges (Fig. 5). An efficient algorithm can be used to remove non-compact pixels by scanning the distance map row wise d − 1 times: A loop is set up to process pixels with a specific distance value dt starting with dt = d − 1 down to 1. The whole distance map is scanned in each cycle. The distance value of pixels pi with di = dt is decremented by 1 if they do not have a paraxial neighbor with a distance value of dt + 1. Finally, pixels pi with di = 0 are removed from the label map. Pseudocode for the non-compact pixel removal can be found as Supplementary Methods S7 online.
(a) 400×400 pixel H&E stained breast cancer tissue, (b) non-overlapping segmentation generated from full contour search (29815 contours in primary segmentation), (c) optimized contours, (d) results after concave object separation and (e) the final segmentation after removing all non-nuclei objects yielding 119 nuclei. The overall segmentation process took 0.39 seconds on a standard PC.
Image object (all pixels) with non-compact pixels on the object border (white pixels).
Numbers represent the distance of the corresponding pixel to the nearest background (non-object) pixel with the Manhattan metric. A distance value d is given for the testing of compactness (3 in this example). Removing all pixels pi (with di the distance value of pi and di < d) that are connected to a pixel pj (with dj = d) over more than d − di edges results in a compact object (gray pixels).
Concave Object Separation (e)
To handle cases of cells forming clusters our approach is based on separating objects at concave borders by removing object pixels (labels) around a cutting line between two concavities (Fig. 4d). Two criteria are taken into account to select the optimal cutting line: The length of the cutting line between two concavities should be minimal compared to the depth of the coressponding concavities and the two corresponding contour concavities should be located opposite to each other. First, concave contour segments are detected in each object (Fig. 6): For that purpose, the convex hull is computed according to36. If at least one contour pixel C exists between two neighboring pixels of the convex hull A and B with distance from AB greater than 0 all contour pixels between A and B are considered concave points and form a concave contour segment. The angle α (clockwise, −π ≤ α ≤ π) of the line segment AB to the x-axis +90° (π/2) is considered the angle of the concavity/concave contour segment. Next, all concave points of all concave contour segments of an object are combined to find the best fitting pair for a separation: This is the cutting line where the so called SeparationScore is minimal (Equation 7). The SeparationScore combines a minimization of the relative cutting line length r (Equation 5) with the “angle criterion” that improves if cutting lines are perpendicular to the convex hull (this helps avoid incorrect cut directions, Equation 6). In Equation (5)–(7) let C1 be the start point, C2 the end point and r the length of the potential cutting line C1C2. depth1 is the distance of C1 from A1B1, depth2 the distance of C2 from A2B2. α1 is the angle between the potential cutting line C1C2 and the corresponding concave contour segment (A1B1) and α2 the angle between the potential cutting line C2C1 and the corresponding concave contour segment (A2B2).



To avoid separations based on concavities with a low depth a minimum depth of concavity may be defined (4 pixels in our examples). Concave borders with a depth below this threshold are ignored in further processing. The separation process is repeated until no further cutting lines are detected. Pseudocode for the concave object separation can be found as Supplementary Methods S8 online.
Classification into cell nuclei and other objects (f)
The classification into cell nuclei and other objects (background) is achieved by using nucleus-specific color information available from histochemical staining. Hematoxylin is a nuclear stain used in routine pathology characterized by a basophilic deep blue/lilac color and is used in combination with immunohistochemical dyes or cytoplasma-specific Eosin stain for conventional histology. To extract the Hematoxylin signal we use color deconvolution (according to37). Subsequently, the staining intensity is computed for each pixel and the optimal threshold is selected from the resulting distribution (according to38). All objects with a mean Hematoxylin staining intensity below this threshold are removed (Fig. 4e). Additionally tiny or artefactual objects with an object area below 50 pixels are removed.
Computational optimizations
For an effective computation the global contour search, contour evaluation, concave object separation and the color deconvolution filter are processed using parallel for loops: Each iteration loop is queued to the Microsoft .NET ThreadPool class. This component assigns, schedules and reuses threads for all queued tasks dependent on the system load and the number of available processors. If background contours (which are removed in the final segmentation step) are already removed in the contour evaluation step these contours do not need to be processed in the subsequent processing steps: Contours with a mean Hematoxylin intensity on contour pixels below the threshold described above are therefore rejected.
Histological sections
We used both whole slides and tissue microarrays derived from tissue samples available through routine diagnostics at the Institute of Pathology, Charité University Medicine Berlin. Patients had given prior consent to their tissue samples being using in scientific studies. The microarrays were constructed by selecting representative tumor areas and then punching out the respective region from formalin-fixed-paraffin-embedded (FFPE) tissue sample blocks and embedded into a new paraffin array block using a tissue microarrayer (Beecher Instruments, Woodland, CA). All sections used were cut 3-μm thick and stained with H&E (Hematoxylin&Eosin) according to standard routine laboratory procedures.
Digitization of histological specimens
The histological slides were digitized using the Zeiss Mirax Scan slide scanner (produced by 3DHistech Ltd, Hungary). The slide scanner was equipped with a Zeiss Plan-Apochromat 20x objective (numerical aperture = 0.8) and an AVT Marlin F-146C Firewire  camera with 4.65 µm x 4.65 µm pixel size. Combined with the 20x objective and 1x C-mount adapter the resulting image resolution is 0.23 µm x 0.23 µm. All slides were scanned at 20x and all processing steps described here were performed at full resolution. No additional components/filters were used. The Zeiss Mirax slide scanner performs an automatic white balance, which corrects the white reference against an area on the glass slide without tissue. The tissue area to be scanned was selected manually. For the scan process we chose to have a focus correction every fourth image tile. The focus is found automatically by the Mirax slide scanning system. Stitching of image tiles was also performed automatically by the Mirax slide scanning system. No other pre-processing was performed. The resulting images were converted to the Virtual Slide Format (VSF) of VMscope GmbH with the actual image data encapsulated and in JPEG format with 85% JPEG quality. The image data is thus compressed, but has the typical JPEG quality and compression used in daily laboratory routine. The file sizes for the whole slide images ranged from 102 MB to 1.96 GB, with an average of 601 MB. The image dimensions ranged from 33280×29184 pixels to 70080×159000 pixels. The analyzed fields of view had a size of 600×600 pixels.
camera with 4.65 µm x 4.65 µm pixel size. Combined with the 20x objective and 1x C-mount adapter the resulting image resolution is 0.23 µm x 0.23 µm. All slides were scanned at 20x and all processing steps described here were performed at full resolution. No additional components/filters were used. The Zeiss Mirax slide scanner performs an automatic white balance, which corrects the white reference against an area on the glass slide without tissue. The tissue area to be scanned was selected manually. For the scan process we chose to have a focus correction every fourth image tile. The focus is found automatically by the Mirax slide scanning system. Stitching of image tiles was also performed automatically by the Mirax slide scanning system. No other pre-processing was performed. The resulting images were converted to the Virtual Slide Format (VSF) of VMscope GmbH with the actual image data encapsulated and in JPEG format with 85% JPEG quality. The image data is thus compressed, but has the typical JPEG quality and compression used in daily laboratory routine. The file sizes for the whole slide images ranged from 102 MB to 1.96 GB, with an average of 601 MB. The image dimensions ranged from 33280×29184 pixels to 70080×159000 pixels. The analyzed fields of view had a size of 600×600 pixels.
Hard- and Software
The image segmentation algorithm was implemented in C# for Microsoft .NET using #Accessory.CognitionMaster39. Virtual Slide Access – SDK 4.0 of VMscope GmbH, Berlin, Germany, was used for the image I/O. All computations were performed on Windows 7 Enterprise x64 SP1 with Intel® Core™ i5-2520M Processor and 4 GB RAM, Barco MDRC-2124 24.1” color LCD-Monitor.
Gold-standard data set preparation and method validation
To assess the accuracy of the proposed method 7931 cells from 36 images were labeled by three pathologists (FK, AS, CD). All images were labeled by three pathologists and only cells for which consensus was achieved were included. All other cells were left unlabeled. The images (600×600 pixels, Hematoxylin-Eosin stained) were taken from previously digitized routine diagnostic cases. After manual labeling the images were segmented with the described method and true positive (tp), false negative (fn) and false positive (fp) events were automatically determined using the positional information (saved in xml-format) previously obtained in the manual “gold-standard” cell labeling. Statistical values were computed as follows:
-
Precision = tp/(tp+fp)
-
Recall = tp/(tp+fn)
-
Conglomerate = (Number of detected cells-fp)/tp
References
Bibbo, M., Bartels, P. H., Dytch, H. E. & Wied, G. L. Computed cell image information. Monogr Clin Cytol 9, 62–100 (1984).
Bengtsson, E. The measuring of cell features. Anal. Quant. Cytol. Histol 9, 212–217 (1987).
Bamford, P. Unsupervised cell nucleus segmentation with active contours. Signal Processing 71, 203–213 (1998).
Bartels, P. H., Gahm, T. & Thompson, D. Automated microscopy in diagnostic histopathology: From image processing to automated reasoning. Int. J. Imaging Syst. Technol 8, 214–223 (1997).
Jiang & Yang An evolutionary tabu search for cell image segmentation. IEEE Trans. Syst. Man, Cybern. B 32, 675–678 (2002).
Latson, L., Sebek, B. & Powell, K. A. Automated cell nuclear segmentation in color images of hematoxylin and eosin-stained breast biopsy. Anal. Quant. Cytol. Histol 25, 321–331 (2003).
Naik, S. et al. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium on, 284–287 (2008).
Wu, Barba, J. & Gil, J. A parametric fitting algorithm for segmentation of cell images. IEEE Trans. Biomed. Eng. 45, 400–407 (1998).
Korde, V. R., Bartels, H., Ranger-Moore, J. & Barton, J. Automatic segmentation of cell nuclei in bladder and skin tissue for karyometric analysis. Anal Quant Cytol Histol. 31, 83–89 (2009).
Tuominen, V. J., Ruotoistenmaki, S., Viitanen, A., Jumppanen, M. & Isola, J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR) and Ki-67. Breast Cancer Res 12, R56 (2010).
Ko, B., Seo, M. & Nam, J.-Y. Microscopic cell nuclei segmentation based on adaptive attention window. J Digit Imaging 22, 259–274 (2009).
Li, G. et al. Segmentation of touching cell nuclei using gradient flow tracking. J Microsc 231, 47–58 (2008).
Reta, C., Gonzalez, J. A., Diaz, R. & Guichard, J. S. Leukocytes segmentation using Markov random fields. Adv. Exp. Med. Biol 696, 345–353 (2011).
Ko, B. C., Gim, J.-W. & Nam, J.-Y. Automatic white blood cell segmentation using stepwise merging rules and gradient vector flow snake. Micron 42, 695–705 (2011).
Bunyak, F., Hafiane, A. & Palaniappan, K. Histopathology tissue segmentation by combining fuzzy clustering with multiphase vector level sets. Adv. Exp. Med. Biol 696, 413–424 (2011).
Wittenberg, T., Grobe, M., Münzenmayer, C., Kuziela, H. & Spinnler, K. A semantic approach to segmentation of overlapping objects. Methods Inf Med 43, 343–353 (2004).
Li, S. Z. Recognizing multiple overlapping objects in image: an optimal formulation. IEEE Trans Image Process 9, 273–277 (2000).
Cristian Smochină. Doctoral Thesis. Technical University “GHEORGHE ASACHI”, 2011.
Yang, L., Tuzel, O., Meer, P. & Foran, D. J. in Lecture Notes in Computer Science, edited by D. Hutchison et al. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2008), pp. 833–841.
Loke, R., Bayer, M., Mann, D. & Du Buf, J. in Oceans '02 MTS/IEEE (IEEE2002), pp. 2457–2465.
INDHUMATHI, C., CAI, Y., GUAN, Y. & OPAS, M. An automatic segmentation algorithm for 3D cell cluster splitting using volumetric confocal images. Journal of Microscopy 243, 60–76 (2011).
Freedman, L. Quantitative science methods for biomarker validation in chemoprevention trials. Cancer Biomark 3, 135–140 (2007).
Sullivan, C. A. W. & Chung, G. G. Biomarker validation: in situ analysis of protein expression using semiquantitative immunohistochemistry-based techniques. Clin Colorectal Cancer 7, 172–177 (2008).
Weigel, M. T. & Dowsett, M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr. Relat. Cancer 17, R245–62 (2010).
Yaziji, H. et al. Consensus recommendations on estrogen receptor testing in breast cancer by immunohistochemistry. Appl. Immunohistochem. Mol. Morphol 16, 513–520 (2008).
Rüschoff, J. et al. HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch 457, 299–307 (2010).
Whenham, N., D'Hondt, V. & Piccart, M. J. HER2-positive breast cancer: from trastuzumab to innovatory anti-HER2 strategies. Clin. Breast Cancer 8, 38–49 (2008).
Underwood, J. C. Nuclear morphology and grading in tumours. Curr Top Pathol 82, 1–15 (1990).
Gerdes, J. Ki-67 and other proliferation markers useful for immunohistological diagnostic and prognostic evaluations in human malignancies. Semin. Cancer Biol 1, 199–206 (1990).
Al-Kofahi, Y., Lassoued, W., Lee, W. & Roysam, B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng 57, 841–852 (2010).
Al-Kofahi, Y. et al. Cell-based quantification of molecular biomarkers in histopathology specimens. Histopathology 59, 40–54 (2011).
Vicidomini, G. et al. High Data Output and Automated 3D Correlative Light-Electron Microscopy Method. Traffic 9, 1828–1838 (2008).
Vicidomini, G. et al. A novel approach for correlative light electron microscopy analysis. Microsc. Res. Tech 73, 215–224 (2010).
Hufnagl, P. & Voss, K. Ein zeitoptimaler Konturfolgealgorithmus (A time-optimal contour search algorithm). “Digitale Bildverarbeitung”, Wiss. Beitr. d. TU Dresden, 18–26 (1983).
BORGEFORS, G. Distance transformations in digital images. Computer Vision, Graphics and Image Processing 34, 344–371 (1986).
Eddy, W. F. A New Convex Hull Algorithm for Planar Sets. ACM Trans. Math. Softw. 3, 398–403 (1977).
Ruifrok, A. C. & Johnston, D. A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol 23, 291–299 (2001).
OTSU N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man, Cybern. 9, 62–66 (1979).
#Accessory.CognitionMaster. http://sourceforge.net/projects/cognitionmaster/.
Acknowledgements
We thank Giuseppe Vicidomini and Carlo Tacchetti (MicroSCoBiO Research Center, University of Genoa, Italy) for providing the confocal fluorescence and electron microscopy data used in Supplementary Note S4. Funding was provided by the Charité Medical School Berlin and the Human Frontier Science Program (HFSP) Young Investigator Grant RGY0077/2011.
Author information
Authors and Affiliations
Contributions
FK, SW, DH and KS wrote the paper. SW and DH prepared the figures. FK, MD, CD, PH and MB planned and supervised the work. SW, DH and FK designed the algorithms. SW and DH implemented the algorithms and performed the data processing and analysis. FK, AS and CD annotated the validation data. FK and AS selected and provided histological samples. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Notes and Methods
Supplementary Information
Supplementary Data S2
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Wienert, S., Heim, D., Saeger, K. et al. Detection and Segmentation of Cell Nuclei in Virtual Microscopy Images: A Minimum-Model Approach. Sci Rep 2, 503 (2012). https://doi.org/10.1038/srep00503
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00503
This article is cited by
-
Real-time and accurate deep learning-based multi-organ nucleus segmentation in histology images
Journal of Real-Time Image Processing (2024)
-
Nuclei probability and centroid map network for nuclei instance segmentation in histology images
Neural Computing and Applications (2023)
-
Detection and Spatiotemporal Analysis of In-vitro 3D Migratory Triple-Negative Breast Cancer Cells
Annals of Biomedical Engineering (2023)
-
Microscopic nuclei classification, segmentation, and detection with improved deep convolutional neural networks (DCNN)
Diagnostic Pathology (2022)
-
Generalising from conventional pipelines using deep learning in high-throughput screening workflows
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.