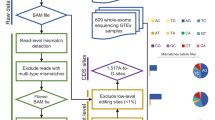
Abstract
We developed a computational framework to robustly identify RNA editing sites using transcriptome and genome deep-sequencing data from the same individual. As compared with previous methods, our approach identified a large number of Alu and non-Alu RNA editing sites with high specificity. We also found that editing of non-Alu sites appears to be dependent on nearby edited Alu sites, possibly through the locally formed double-stranded RNA structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Rosenberg, B.R., Hamilton, C.E., Mwangi, M.M., Dewell, S. & Papavasiliou, F.N. Nat. Struct. Mol. Biol. 18, 230–236 (2011).
Nishikura, K. Annu. Rev. Biochem. 79, 321–349 (2010).
Levanon, E.Y. et al. Nat. Biotechnol. 22, 1001–1005 (2004).
Silberberg, G. & Ohman, M. Curr. Opin. Genet. Dev. 21, 401–406 (2011).
Li, J.B. et al. Science 324, 1210–1213 (2009).
Li, M. et al. Science 333, 53–58 (2011).
Ju, Y.S. et al. Nat. Genet. 43, 745–752 (2011).
Bahn, J.H. et al. Genome Res. 22, 142–150 (2012).
Peng, Z. et al. Nat. Biotechnol. 30, 253–260 (2012).
Schrider, D.R., Gout, J.F. & Hahn, M.W. PLoS One 6, e25842 (2011).
Lin, W., Piskol, R., Tan, M.H. & Li, J.B. Science 335, 1302 (2012).
Kleinman, C.L. & Majewski, J. Science 335, 1302 (2012).
Pickrell, J.K., Gilad, Y. & Pritchard, J.K. Science 335, 1302 (2012).
Li, H. & Durbin, R. Bioinformatics 25, 1754–1760 (2009).
Neeman, Y., Levanon, E.Y., Jantsch, M.F. & Eisenberg, E. RNA 12, 1802–1809 (2006).
Bustamante, C.D., Burchard, E.G. & De la Vega, F.M. Nature 475, 163–165 (2011).
Kiran, A. & Baranov, P.V. Bioinformatics 26, 1772–1776 (2010).
Parkhomchuk, D. et al. Nucleic Acids Res. 37, e123 (2009).
Li, H. & Homer, N. Brief. Bioinform. 11, 473–483 (2010).
Li, H. et al. Bioinformatics 25, 2078–2079 (2009).
Durbin, R.M. et al. Nature 467, 1061–1073 (2010).
Acknowledgements
We thank E. Levanon and N. Sanjana for critical reading of the manuscript, C. Pan and J. Sun for technical assistance, and T. Gingeras for support. This work is supported by the Stanford University Department of Genetics and the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
G.R., W.L. and R.P. performed the computational analyses with help from M.H.T. and J.B.L. M.H.T. and G.R. carried out the validation experiments. C.D. generated the GM12878 RNA-seq data. R.P. and J.B.L. wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 1–5, and Supplementary Notes 1–4 (PDF 2511 kb)
Supplementary Data 1
All RNA editing sites identified in GM12878 using our method. (XLSX 8161 kb)
Supplementary Data 2
All RNA editing sites identified in YH using our method. (XLSX 47856 kb)
Rights and permissions
About this article
Cite this article
Ramaswami, G., Lin, W., Piskol, R. et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods 9, 579–581 (2012). https://doi.org/10.1038/nmeth.1982
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1982
This article is cited by
-
Deep transcriptome profiling reveals limited conservation of A-to-I RNA editing in Xenopus
BMC Biology (2023)
-
DeepEdit: single-molecule detection and phasing of A-to-I RNA editing events using nanopore direct RNA sequencing
Genome Biology (2023)
-
The epitranscriptome of high-grade gliomas: a promising therapeutic target with implications from the tumor microenvironment to endogenous retroviruses
Journal of Translational Medicine (2023)
-
L-GIREMI uncovers RNA editing sites in long-read RNA-seq
Genome Biology (2023)
-
Transcriptome-wide profiling and quantification of N6-methyladenosine by enzyme-assisted adenosine deamination
Nature Biotechnology (2023)