« Prev Next »
Types of Transposons
Today, scientists know that there are many different types of TEs, as well as a number of ways to categorize them. One of the more common divisions is between those TEs that require reverse transcription (i.e., the transcription of RNA into DNA) in order to transpose and those that do not. The former elements are known as retrotransposons or class 1 TEs, whereas the latter are known as DNA transposons or class 2 TEs. The Ac/Ds system that McClintock discovered falls in the latter category. Different classes of transposable elements are found in the genomes of different eukaryotic organisms (Figure 1).
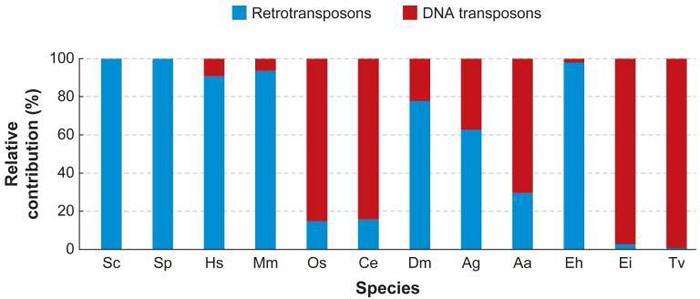
DNA Transposons
All complete or "autonomous" class 2 TEs encode the protein transposase, which they require for insertion and excision (Figure 2). Some of these TEs also encode other proteins. Note that DNA transposons never use RNA intermediaries—they always move on their own, inserting or excising themselves from the genome by means of a so-called "cut and paste" mechanism.

Class 2 TEs are characterized by the presence of terminal inverted repeats, about 9 to 40 base pairs long, on both of their ends (Figure 3). As the name suggests and as Figure 3 shows, terminal inverted repeats are inverted complements of each other; for instance, the complement of ACGCTA (the inverted repeat on the right side of the TE in the figure) is TGCGAT (which is the reverse order of the terminal inverted repeat on the left side of the TE in the figure). One of the roles of terminal inverted repeats is to be recognized by transposase.
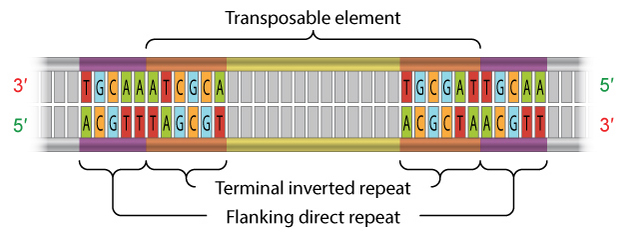
In addition, all TEs in both class 1 and class 2 contain flanking direct repeats (Figure 3). Flanking direct repeats are not actually part of the transposable element; rather, they play a role in insertion of the TE. Moreover, after a TE is excised, these repeats are left behind as "footprints." Sometimes, these footprints alter gene expression (i.e., expression of the gene in which they have been left behind) even after their related TE has moved to another location on the genome.
Less than 2% of the human genome is composed of class 2 TEs. This means that the majority of the substantial portion of the human genome that is mobile consists of the other major class of TEs—the retrotransposons (Kazazian & Moran, 1998).
Retrotransposons
There are two major types of class 1 TEs: LTR retrotransposons, which are characterized by the presence of long terminal repeats (LTRs) on both ends; and non-LTR TEs, which lack the repeats. Both the LINE1, or L1, and Alu genes represent families of non-LTR TEs. L1 elements average about 6 kilobases in length. In contrast, Alu elements average only a few hundred nucleotides, thus making them a short interspersed transposable element, or SINE. Alu is particularly prolific, having originated in primates and expanding in a relatively short time to about 1 million copies per cell in humans. L1 is also common in humans; although not present in as many copies as Alu, its larger size means that this element makes up an estimated 15%–17% of the human genome (Kazazian & Moran, 1998; Slotkin & Martienssen, 2007). In humans, these non-LTR TEs are the only active class of transposons; LTR retrotransposons and DNA transposons are only ancient genomic relics and are not capable of jumping.
Autonomous and Nonautonomous Transposons
What Jumping Genes Do (Besides Jump)
The fact that roughly half of the human genome is made up of TEs, with a significant portion of them being L1 and Alu retrotransposons, raises an important question: What do all these jumping genes do, besides jump? Much of what a transposon does depends on where it lands. Landing inside a gene can result in a mutation, as was discovered when insertions of L1 into the factor VIII gene caused hemophilia (Kazazian et al., 1988). Similarly, a few years later, researchers found L1 in the APC genes in colon cancer cells but not in the APC genes in healthy cells in the same individuals. This confirms that L1 transposes in somatic cells in mammals, and that this element might play a causal role in disease development (Miki et al., 1992).
Silencing and Transposons
Another example of transposon silencing involves plants in the genus Arabidopsis. Researchers who study these plants have found they contain more than 20 different mutator transposon sequences (a type of transposon identified in maize). In wild-type plants, these sequences are methylated, or silenced. However, in plants that are defective for one of the enzymes responsible for methylation, these transposons are transcribed. Moreover, several different mutant phenotypes have been explored in the methylation-deficient plants, and these phenotypes have been linked to transposon insertions (Miura et al., 2001).
Based on studies such as these, scientists know that some TEs are epigenetically silenced; in recent years, however, researchers have begun to wonder whether certain TEs might themselves have a role in epigenetic silencing. Interestingly, it was Barbara McClintock who first speculated that TEs might play this kind of regulatory role (McClintock, 1951). It has taken decades for scientists to collect enough evidence to consider that maybe McClintock's speculation had a kernel of truth to it.
Transposons Can Encode siRNAs That Mediate Their Own Silencing
For instance, in human cells, small interfering RNAs (siRNAs), also known as RNAi, can prevent transposition. RNAi is a naturally occurring mechanism that eukaryotes often use to regulate gene expression. What is especially interesting about this situation is that the siRNAs that interfere with L1 activity are derived from the 5′ untranslated region (5′ UTR) of L1 LTRs. Specifically, the 5′ UTR of the L1 promoter encodes a sense promoter that transcribes the L1 genes, as well as an antisense promoter that transcribes an antisense RNA. Yang and Kazazian (2006) demonstrated that this results in homologous sequences that can hybridize, thereby forming a double-stranded RNA molecule that can serve as a substrate for RNAi. Furthermore, when the investigators inhibited endogenous siRNA silencing mechanisms, they saw an increase in L1 transcripts, suggesting that transcription from L1 is indeed inhibited by siRNA.
Transposons Are Not Always Destructive
The fact that transposable elements do not always excise perfectly and can take genomic sequences along for the ride has also resulted in a phenomenon scientists call exon shuffling. Exon shuffling results in the juxtaposition of two previously unrelated exons, usually by transposition, thereby potentially creating novel gene products (Moran et al., 1999).
The ability of transposons to increase genetic diversity, together with the ability of the genome to inhibit most TE activity, results in a balance that makes transposable elements an important part of evolution and gene regulation in all organisms that carry these sequences.
References and Recommended Reading
Feschotte, C., et al. Plant transposable elements: Where genetics meets genomics. Nature Reviews Genetics 3, 329–341 (2002) (link to article)
Kazazian, H. H. Mobile elements: Drivers of genome evolution. Science 303, 1626–1632 (2004) doi:10.1126/science.1089670
Kazazian, H. H., & Moran, J. V. The impact of L1 retrotransposons on the human genome. Nature Genetics 19, 19–24 (1998) (link to article)
Kazazian, H. H., et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166 (1988) (link to article)
Koga, A., et al. Vertebrate DNA transposon as a natural mutator: The medaka fish Tol2 element contributes to genetic variation without recognizable traces. Molecular Biology and Evolution 23, 1414–1419 (2006) doi:10.1093/molbev/msl003
McLean, P. McClintock and the Ac/Ds transposable elements of corn, www.ndsu.nodak.edu/instruct/mcclean/plsc431/transelem/trans1.htm (1997)
McClintock, B. Mutable loci in maize. Carnegie Institution of Washington Yearbook 50, 174–181 (1951) (link to article)
Miki, Y., et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in colon cancer. Cancer Research 52, 643–645 (1992)
Miura, A., et al. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214 (2001) (link to article)
Moran, J. V., et al. Exon shuffling by L1 retrotransposition. Science 283, 1530–1534 (1999)
SanMiguel, P., et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768 (1996)
Slotkin, R. K., & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics 8, 272–285 (2007) (link to article)
Yang, N., & Kazazian, H. H. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature Structural and Molecular Biology 13, 763–771 (2006) (link to article)






























