« Prev Next »
Transposable elements (TEs), also known as "jumping genes" or transposons, are sequences of DNA that move (or jump) from one location in the genome to another. Maize geneticist Barbara McClintock discovered TEs in the 1940s, and for decades thereafter, most scientists dismissed transposons as useless or "junk" DNA. McClintock, however, was among the first researchers to suggest that these mysterious mobile elements of the genome might play some kind of regulatory role, determining which genes are turned on and when this activation takes place (McClintock, 1965).
At about the same time that McClintock performed her groundbreaking research, scientists Roy Britten and Eric Davidson further speculated that TEs not only play a role in regulating gene expression, but also in generating different cell types and different biological structures, based on where in the genome they insert themselves (Britten & Davidson, 1969). Britten and Davidson hypothesized that this might partially explain why a multicellular organism has many different types of cells, tissues, and organs, even though all of its cells share the same genome. Consider your own body as an example: You have dozens of different cell types, even though the majority of cells in your body have exactly the same DNA. If every single gene was expressed in every single one of your cells all the time, you would be one huge undifferentiated blob of matter!
The early speculations of both McClintock and Britten and Davidson were largely dismissed by the scientific community. Only recently have biologists begun to entertain the possibility that this so-called "junk" DNA might not be junk after all. In fact, scientists now believe that TEs make up more than 40% of the human genome (Smit, 1999). It is also widely believed that TEs might carry out some biological function, most likely a regulatory one—just as McClintock and Britten and Davidson speculated. Like all scientific hypotheses, however, data from multiple experiments were required to convince the scientific community of this possibility.
The SINE Superfamily of Transposons
Much of the evidence for the function of TEs comes from the growing realization that many transposons are highly conserved among distantly related taxonomic groups, suggesting that they must be of some biological value to the genome (Pennisi, 2007). To say that a DNA sequence is conserved means that the same TEs, or a family of related TEs, exist in genomes of distantly related species, such as fish and frogs.
For instance, in one study, Japanese researchers identified a new "superfamily" of TEs (a group of TEs that share some similarities because of their shared ancestry), which they dubbed V-SINEs. The scientists then used PCR to show that the V-SINEs were widespread among vertebrates, including lampreys, cartilaginous fish, bony fish, and amphibians (Ogiwara et al., 2002). Of these vertebrates, lampreys are the oldest, having emerged in the Cambrian era (544 to 510 million years ago). As the scientists explained in their paper:
The distribution of V-SINEs suggests that they might have been generated in a common ancestor of vertebrates and might then have survived in most vertebrates . . . [and] that they might have been generated in the genome of a common ancestor of vertebrates about 540 million years ago or even more and might then have survived in most vertebrates until the present day.
These findings suggest that V-SINEs are approximately 540 million years old.
V-SINEs acquired their name from the SINEs, or short interspersed nuclear elements. SINEs are a type of non-long terminal repeat retrotransposon (non-LTR TE). Like all non-LTR TEs, SINEs (including V-SINEs) are retrotransposons; this means that their movement around the genome is dependent on the presence of an RNA intermediary. In this system, the TEs produce RNA transcripts, which are then converted back into DNA by an enzyme called reverse transcriptase. The new DNA copies then insert themselves into other spots in the genome.
While the researchers who discovered V-SINEs speculate on their function—specifically, whether they have something to do with protein production during times of stress, based on the fact that other researchers have observed greater SINE transcription under stressful conditions—they emphasize that the true functions of V-SINEs remain a scientific mystery (Ogiwara et al., 2002). The fact that the V-SINEs are so highly conserved suggests that, even though their true function is still unknown, these TEs must have some specific role. If not, then why do so many different species share the same, or similar, V-SINEs? Over 500 million years is a long time for "junk" to survive if it has no purpose.
Conserved SINE Sequences
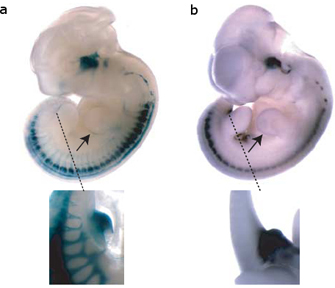
 Figure 1: The SINE transposable element upstream of the ISL1 gene serves to enhance gene expression.
Figure 1: The SINE transposable element upstream of the ISL1 gene serves to enhance gene expression.
Perhaps even more intriguingly, Bejerano and his colleagues collected data suggesting that the LF-SINE family TEs likely play a regulatory role, just as McClintock, Britten, and Davidson suspected decades ago. First, Bejerano et al. identified a 200-base-pair LF-SINE that resided some 500,000 bases away from the Isl1 gene, which is active only during motor neuron development. Then, using a procedure called a mouse enhancer assay, the researchers showed that this TE controlled expression of Isl1—specifically, only when the TE was turned on was Isl1 turned on. The mouse enhancer assay involved linking the TE sequence to a gene that would cause cells within a developing mouse embryo to turn blue when expressed, making the cells readily identifiable (Figure 1). The expression patterns associated with the color change (and therefore associated with activity of the TE) corresponded to Isl1 expression changes typical of a particular developmental stage, suggesting that the TE functions as an enhancer, or a regulatory element that can activate a gene from a distance.
Later researchers have found similar results—that TEs can influence gene transcription—in other species, such as fruit flies, morning glory flowers, and (vindicating McClintock's suspicions) maize (Slotkin & Martienssen, 2007). Moreover, in primates, scientists have identified a SINE known as Alu that seems to play an important role in gene regulation and evolution. These new discoveries are prompting scientists to think twice about dismissing such a large portion of the genome as nothing but "junk."
References and Recommended Reading
Bejerano, G., et al. A distal enhancer and an ultra conserved exon are derived from a novel retrotransposon. Nature 441, 87–90 (2006) doi:10.1038/nature04696 (link to articles)
Britten, R. J., & Davidson, E. H. Gene regulation for higher cells: A theory. Science 165, 349–357 (1969)
McClintock, B. Components of action of the regulators Spm and Ac. Carnegie Institution of Washington Year Book 64, 527–536 (1965)
Ogiwara, I., et al. V-SINEs: A new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Research 12, 316–324 (2002)
Pennisi, E. Jumping genes hop into the evolutionary limelight. Science 317, 894–895 (2007)
Slotkin, R. K., & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nature Reviews Genetics 8, 272–285 (2007) doi:10.1038/nrg2072 (link to articles)
Smit, A. F. A. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Current Opinion in Genetics and Development 9, 657–663 (1999)






























