« Prev Next »
A diverse range of microbes, including viruses, bacteria, fungi, and protists, stand ready to attack the human body and thrive in the nutrient-rich environment it provides. Fortunately, the immune response functions as a defense mechanism and counterattacks by recognizing and destroying foreign invaders. But what alerts the body to danger? How are foreign organisms detected? The discovery of microbial-sensing proteins called Toll-like receptors is helping to answer these questions and transform our understanding of the response to infection.
What Do Toll-Like Receptors Do?
A small number of Toll-like receptors can detect a broad range of human pathogens, as well as a variety of other molecules that indicate tissue damage, by a process called pattern recognition. These receptors initiate two arms of the immune response — the innate and adaptive responses — that work together to fight infection in mammals. The innate response provides immediate protection. However, it is relatively nonspecific in its mode of attack on pathogens, which results in damage to healthy tissue if the innate immune response lasts too long. The adaptive response, on the other hand, generates antibody-secreting B cells and cytotoxic T cells that are specific and efficient at targeting pathogen. Unfortunately, this process takes longer to develop than the innate response.
Because Toll-like receptors function as first responders to danger signals, they are centrally significant in research efforts to combat infectious and inflammatory disease. New strategies for manipulating immune responses depend on understanding the cell biology of Toll-like receptors, including their structure, cell localization, signal transduction pathways, and expression patterns.
Proteins, Receptors, and Environment: The Basics
The problem of microbial detection is part of a more general problem in cell biology: How can a cell obtain information about its environment? Even single-celled organisms must monitor their surroundings and respond to external signals to survive. Our bodies are multicellular, and large numbers of cells need to communicate with one another and coordinate their activities to function as an integrated whole. The boundary of each cell is defined by the phospholipid bilayer of the plasma membrane, which acts as an effective barrier to most water-soluble molecules. This boundary is not inert. The inclusion of proteins in the phospholipid bilayer makes the membrane a dynamic entity capable of sensing and interacting with the cellular environment. For example, membrane receptors bind and respond to specific molecules outside the cell, such as hormones, neurotransmitters, the extracellular matrix, and attachment molecules on other cells.
Proteins perform a wide range of biological functions because of their versatile structure. Linear chains of amino acids form proteins, and protein diversity arises from different amino acid combinations. Just as the twenty-six letters of the alphabet create a wide variety of words when combined, the twenty different amino acids create a variety of proteins when combined. Each protein has a unique amino acid sequence that determines how the chain folds into a three-dimensional structure. The result is the tertiary structure or conformation of the protein, and it serves a unique, particular function. A receptor has a three-dimensional shape that enables it to bind with another molecule called a ligand (which might be a hormone, a neurotransmitter, etc.). The receptor and ligand bind with high affinity, meaning that the interaction is very strong, and with high specificity, meaning that binding is exclusive to the ligand or very similar molecules.
Pattern Recognition and Toll
Human cells have only about 25,000 protein-encoding genes, so it is impossible to have a different gene (and a different receptor) for each species of virus, bacteria, protist, and fungus. How, then, can the body identify all species of pathogens that pose a danger, even those it has never encountered before? In 1989, Charles Janeway proposed that cells use pattern recognition to detect pathogens (Janeway 1989). In other words, receptors bind to structural shapes or patterns called PAMPs (pathogen-associated molecular patterns) that are present in whole groups of pathogens, but not the host. According to Janeway's theory, receptors cannot identify a particular microbe with precision, but they can recognize it as a foreign organism.
The first human pattern-recognition receptors were identified ten years after Janeway's proposal. The breakthrough was made possible by an earlier discovery in the fruit fly Drosophila. For decades, researchers had used Drosophila to identify developmental mutations, but Drosophila was not considered a good choice to model human immunity due to the lack of an adaptive response in insects (Beck & Habicht 1996). German scientists originally identified the Toll gene as the site of mutations that generated bizarre-looking flies. (They exclaimed that their results were "Toll!" which transIates to "Great!" in English.) Cloning the gene demonstrated that it encoded a membrane receptor (Hashimoto, Hudson & Anderson 1988).
A 1996 study reported that loss-of-function Toll mutations made Drosophila highly susceptible to fungal infections and that gain-of-function mutations led to increased production of certain antifungal proteins (Lemaitre et al. 1996). Comparisons of Toll mutations to mutations in other genes led to the conclusion that the Toll receptor plays a dominant role in detecting fungal infections and initiating the innate immune response. This exciting discovery provided researchers with the clue they needed to find human pathogen receptors. Using the amino acid sequence of Toll, they searched for related sequences in the Human Genome Project database and identified Toll-like receptors (Medzhitov, Preston-Hurlburt & Janeway 1997; Rock et al. 1998).
Understanding Toll-Like Receptors
Mouse models are used to study a number of human inflammatory diseases, which are caused by an overactive innate immune response that leads to dangerous inflammation and tissue destruction. For example, sepsis is a severe illness in which the bloodstream is overwhelmed with bacteria, causing inflammation throughout the body. It is responsible for the deaths of more than 200,000 people in the United States each year. Creating mouse models of sepsis involves injecting mice with bacteria to induce inflammation. The severity of disease is then measured with controlled and standardized assays. Scientists have known for many years that isolated bacterial components, such as lipopolysaccharide (LPS), can substitute for whole bacteria to induce sepsis. LPS is present in the cell walls of all Gram-negative bacteria, which represents a large group of pathogens. It is, therefore, an example of Janeway's PAMPs. In addition, Poltorak et al. (1998) demonstrated that mice with a defective version of TLR4 are completely resistant to LPS-induced sepsis. This established that TLR4 is the receptor for LPS, and that it generates an innate immune response following LPS
stimulation.
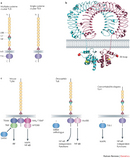
Scientists now know that humans have at least ten different TLRs, and they collectively recognize a broad spectrum of pathogens (Figure 1). TLRs 1, 2, 4, 5, and 6 bind to components of microbial cell walls and membranes unique to pathogens. The best characterized ligands are bacterial, including LPS and lipoteichoic acid from cell walls, lipoproteins from the cell membrane, and a structural component of bacterial flagella called flagellin. TLRs 3, 7, 8, and 9 bind to microbial nucleic acids, including double and single-stranded RNA from RNA viruses and DNA from most organisms. These TLRs cannot distinguish self-nucleic acids (those of the host cell) on structural differences alone, and recognition of foreign nucleic acids (those of the pathogen) largely depends on the location in the cell. All these ligands play an essential role in the microbe, and the microbe cannot eliminate or modify them to evade detection. Scientists have found TLRs in lower animals, such as the nematode C. elegans, and in plants, indicating that TLRs have an ancient evolutionary origin in host defense. Other pattern-recognition receptors for pathogens are also being identified (Figure 1).
TLR Structure
Scientists have deduced the three-dimensional structure of TLRs with x-ray crystallography (example, Figure 2b). The analysis of protein tertiary structure is a complicated puzzle, and figuring out the structure of something like a TLR is always a significant accomplishment. For TLRs, the precise molecular details reveal how ligand-receptor interactions emerge from the properties of atoms within the receptor (Jin et al. 2007; Liu et al. 2008; Park et al. 2009). This information explains the specificity of a receptor and permits design of chemical agonists, which mimic the ligand's function, or chemical antagonists, which block the ligand's action. For example, chemists can use the molecular structure of TLR4 and its interaction with LPS to infer the best design for TLR4 antagonists, which can be treatments for certain inflammatory disorders, including sepsis (Park et al. 2009).
TLR Localization and Cell-Specific Expression
TLRs 1, 2, 4, 5, and 6 are located primarily in the plasma membrane, where they interact with components of microbial pathogens that come into contact with the cell. In contrast, TLRs 3, 7, 8, and 9 are situated in the membranes of endosomes and lysosomes; the extracellular domain of the receptor and its ligand-binding site project into the interior of these organelles. After these organelles break down pathogens that have been internalized by endocytosis, TLRs can interact with the now-exposed pathogen DNA and RNA.
TLRs have complex expression patterns in different cell types. Recall that different cell types of a multicellular organism share the same set of genes, but specialization of cell function is possible because different genes are expressed. TLR expression is particularly significant in different types of white blood cells: mast cells, macrophages, and dendritic cells. These cells leave the bloodstream and take up residence in tissues throughout the body, where they are detectors of pathogens. Innate immune responses are initiated by mast cells and macrophages, whereas adaptive immune responses are primarily initiated by dendritic cells.
Damage Signals
There is an unexpected twist to the story of TLRs. Most scientists assumed that TLR function is specialized to detect foreign organisms so that the body can distinguish self from nonself. However, there is now convincing evidence that some TLR ligands originate from the host. Why would the immune system react to self-components? The hypothesis is that these new ligands act as damage signals (damage-associated molecular patterns, or DAMPs) to alert the body of cell and tissue injury. Many injuries result in a messy form of cell death called necrosis, which releases intracellular components into the surroundings, and some of these components specifically activate TLRs (Yang et al. 2010). Breakdown products of the extracellular matrix may also act as damage signals (Schaefer 2010). Scientists are still speculating on the purpose of these endogenous signals. They might initiate tissue repair responses, initiate immune responses as a preemptive strike against infections that usually follow injury, or collaborate with TLRs to identify pathogens that cause injury and distinguish them from harmless commensal microbes.
Endogenous damage signals can stimulate an inappropriate innate immune response that does more harm than good in some cases in which there is no infection. An example is ischemic injury that results from the temporary loss of blood flow and hence oxygen supply. Ischemic injury is a concern in many clinical scenarios, including stroke and transplantation. Cell death during ischemia releases damage signals, and restoration of blood flow allows an influx of white blood cells, which cause inflammation in response to the signals. This inflammation leads to more destruction than the original injury. Blocking TLR2 and TLR4 with antagonists may be useful in these circumstances to prevent an overactive immune response (Gill, Tsung & Billiar 2010). There is also evidence that TLRs contribute to the development of atherosclerosis and Alzheimer's disease through sensing of damage signals in the form of oxidized lipoproteins (Stewart et al. 2010).
TLR Signal Transduction
After TLRs detect pathogen or damage-associated molecular patterns, they must convert these signals into messages inside a cell via signal transduction (Figure 2c). Often, it is these signaling events that scientists want to change when they consider designing drugs for disease intervention. Signaling components inside a cell called protein kinases activate other proteins by adding a phosphate group to certain amino acids, and they are among the best characterized drug targets. Approximately ten protein kinase inhibitors have already been approved for the treatment of cancer. TLR signaling involves a variety of protein kinases, including IRAKs (IL-1R-associated kinases) and MAPKs (mitogen-activated protein kinases), and some of them are promising targets for drug development. For example, mice are resistant to septic shock and chronic inflammation if they have a defective form of IRAK4 (Cohen 2009); thus, inhibitors of IRAK4 may help prevent septic shock and inflammation in humans. The final response of a cell to TLR stimulation usually involves activation of transcription factors that regulate specific patterns of gene expression, leading to an increased production of hundreds of proteins. They include small proteins called cytokines, which are secreted from the cell to recruit and activate other cells, thus initiating and orchestrating the wider immune response.
Future Prospects
The discovery of TLRs and their cell biology provides new opportunities for drug intervention to manipulate immune responses (Hennessy, Parker, & O'Neill 2010). TLRs are most clearly associated with initiation of the innate response and inflammation (Takeuchi & Akira 2010), and inhibition of TLR activity may help combat an overactive innate response characteristic of numerous inflammatory disorders.
The roles of TLRs in the adaptive immune response are less well understood, and this subject is an important area of future study (Iwasaki & Medzhitov 2010). Vaccines for diseases such as AIDS, hepatitis C, malaria, and even cancer might be made more effective by supplementing them with TLR activators that stimulate dendritic cells. The activated dendritic cells then produce a more effective adaptive immune response. For example, scientists have linked a conserved component of influenza viruses with an agonist to TLR5 to create a vaccine that could potentially protect against all strains of flu (Huleatt et al. 2008).
References and Recommended Reading
Beck, G. & Habicht, G. S. Immunity and the invertebrates. Scientific American 275, 60–66 (1996).
Blasius, A. L. & Beutler, B. Intracellular Toll-like receptors. Immunity 32: 305–315 (2010). doi: 10.1016/j.immuni.2010.03.012
Cohen, P. Targeting protein kinases for the development of anti-inflammatory drugs. Current Opinion in Cell Biology 21, 317–324 (2009) doi: 10.1016/j.ceb.2009.01.015.
Gill, R., Tsung, A. & Billiar, T. Linking oxidative stress to inflammation: Toll-like receptors. Free Radical Biology and Medicine 48, 1121–1132 (2010) doi:10.1016/j.freeradbiomed.2010.01.006.
Hashimoto, C., Hudson, K. L. & Anderson, K. V. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell 52, 269–279 (1988) doi:10.1016/0092-8674(88)90516-8.
Hennessy, E. J., Parker, A. E. & O'Neill, A. J. Targeting of Toll-like receptors: Emerging therapeutics? Nature Reviews Drug Discovery 9, 293–307 (2010) doi: 10.1038/nrd3203.
Huleatt, J. W. et al. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 26, 201–214 (2008) doi:10.1016/j.vaccine.2007.10.062.
Iwasaki, A. & Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 327, 291–295 (2010) doi:10.1126/science.1183021.
Janeway, C. A. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symposia on Quantitative Biology 54, 1–13 (1989).
Jin, M. S. et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 (2007) doi:10.1016/cell.2007.09.008.
Lemaitre, B. et al. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996) doi:10.1016/S0092-8674(00)80172-5.
Leulier, F. & Lemaitre, B. Toll-like receptors — taking an evolutionary approach. Nature Reviews Genetics 9, 165–178 (2008) doi:10.1038/nrg2303.
Liu, L. et al. Structural basis of Toll-like receptor 3 signaling with double stranded RNA. Science 320, 379–381 (2008) doi:10.1126/science.1155406.
Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997).
Park, B. S. et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458 1191–1195 (2009) doi:10.1038/nature07830.
Poltorak, A. et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282: 2085-
2088 (1998) doi: 10.1006/bcmd.1998.0201.
Rock, F. L. et al. A family of human receptors structurally related to Drosophila Toll. Proceedings of the National Academy of Sciences 95, 588–593 (1998).
Schaefer, L. Extracellular matrix molecules: Endogenous danger signals as new drug targets in kidney diseases. Current Opinion in Pharmacology 10, 185–190 (2009) doi:10.1016/j.coph.2009.11.007.
Stewart, C. R. et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature Immunology 11,155–161 (2010) doi:10.1038/ni.1836.
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010) doi:10.1016/j.cell.2010.01.022.
Yang, H. et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proceedings of the National Academy of Sciences 107, 11942–11947 (2010) doi:10.1073/pnas.1003893107.



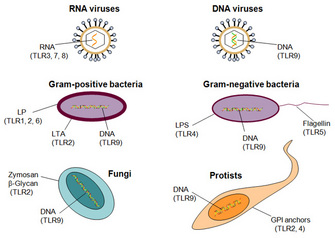
 Figure 1: Receptors that detect infection
Figure 1: Receptors that detect infection