« Prev Next »
Scientists have determined the atomic structures of thousands of the biomolecular components of cells. These structures allow us to understand cell biology at the atomic level. The secrets of protein synthesis have been revealed, starting with the structure of DNA half a century ago, revealing the atomic basis of genetic information, to the recent structures of working ribosomes, caught in the act of translating this information into new proteins. The enzymes of glycolysis, citric acid cycle, and electron transport have all been studied and their structures have been determined. Three-dimensional (3D) structures are known for diverse proteins of sense, signaling, transport, regulation, and defense. These structures answer many important biological questions, and also allow scientists to pose many new ones.
Most Biomolecular Structures Are Determined by X-ray Crystallography
How do scientists determine the atomic structures of proteins? X-ray crystallography is a powerful method for determining the location of every atom in a molecule. The biomolecule of interest is purified and crystallized, and then the crystal is subjected to an intense beam of X-rays (often, these days, provided by a synchotron). The X-rays are diffracted into a characteristic array of spots. By analyzing the spacing of the spots, researchers can determine how the molecules are arranged in the crystal, and by analyzing the intensity of the spots, they can determine where each atom lies. The result is a set of coordinates, describing the location of each atom in the molecule.
Other Methods Are Also Used to Determine Structures
Scientists use a variety of other techniques to determine the three-dimensional structures of biomolecules, particularly for molecules that are too flexible or too large to be analyzed by crystallography. NMR spectroscopy reveals all of the atoms that are close to one another in a biomolecule, as well as the local conformation of portions of the macromolecular chain. Scientists then use this information to infer the structure of the biomolecule. Currently, NMR spectroscopy is effective for determining the structures of small biomolecules, such as small proteins or oligonucleotides, since the experimental measurements are difficult or impossible to make for larger molecules. Electron microscopy, on the other hand, is effective for very large structures, such as large viruses or assemblies of proteins like the nuclear pore. The optics used to focus electrons are not quite accurate enough to resolve individual atoms, but electron microscopy can typically give a good indication of the overall shape and form of subcellular molecules.
Biomolecular Structures Are Freely Available
What do scientists do with all the information about molecular structure? Structural biology is one of the earliest fields of biology to make the primary research data directly available to the general public. These atomic structures have been available from the Protein Data Bank archive (PDB) since 1971 (Bernstein et al. 1977). The PDB is the central repository of biomolecular structures, and researchers in structural biology typically deposit their coordinates soon after the completion of their study. After deposition, the data are reviewed, annotated, and made freely available by a consortium of data centers called the Worldwide PDB (Berman, Henrick, and Nakamura 2003). The research community has determined that public access is critical for the advancement of science, so most journals and granting agencies have strict requirements for deposition of coordinates. Currently, there are more than 65,000 entries of biomolecules available at the PDB, solved and deposited by researchers around the globe.
Biomolecular Structures Are Used to Discover New Drugs
Why is knowing the atomic structure of molecules useful? Coordinate data files representing proteins and nucleic acids are freely available online, and are actually used for many research projects. One of the most important applications of biomolecular structure is drug design. When we know the structure of a protein, we can attempt to design small drug molecules to bind to it and block its function. The power of this approach has been shown in the battle against HIV and AIDS. Many of the anti-HIV drugs that are currently saving lives were discovered with the help of knowing the structures of HIV proteins (Figure 1).
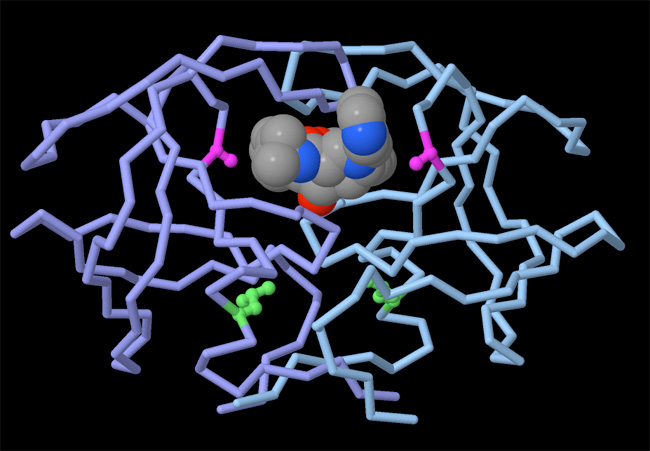
Biomolecular Structures Reveal the Atomic Details of Life
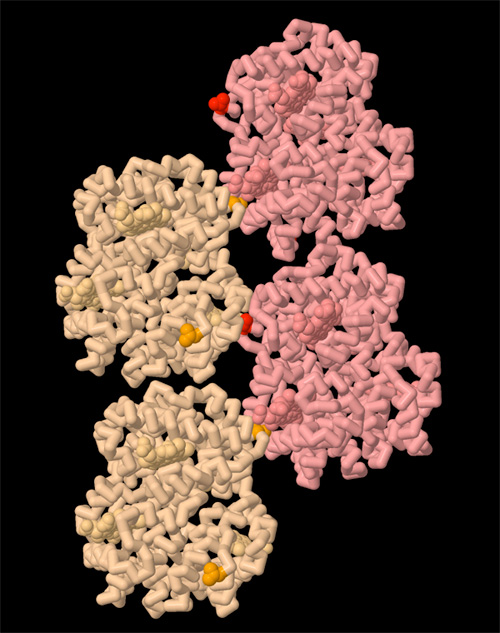
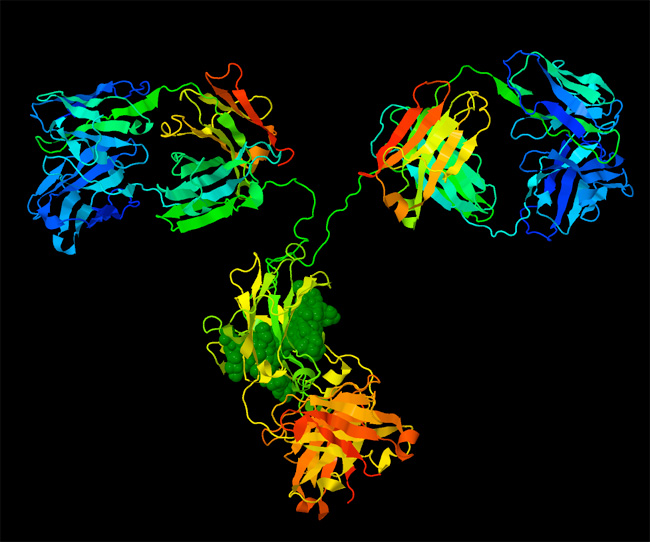
Biomolecular structures have also been instrumental in understanding the basic mechanisms that keep cells alive. For instance, structures of oxy and deoxy hemoglobin revealed the atomic basis of allostery for control of oxygen binding, and structures of sickle cell hemoglobin revealed the atomic basis of a disease-causing mutation (Figure 2). With an atomic structure, it has been possible to explore the detailed mechanism of enzymes, and understand how they stabilize transition states. Atomic structures have revealed the complex motions of motor proteins, including the flexing of myosin from our muscles and the linked rotary motors of ATP synthase. In addition, atomic structures have revealed the basis of immune system function, as they have shown how antibodies recognize foreign molecules, and how MHC signals viral infection. Each new structure adds a new piece in the puzzle of how life works.
Scientists Design New Molecular Machines Based on Biomolecular Structures
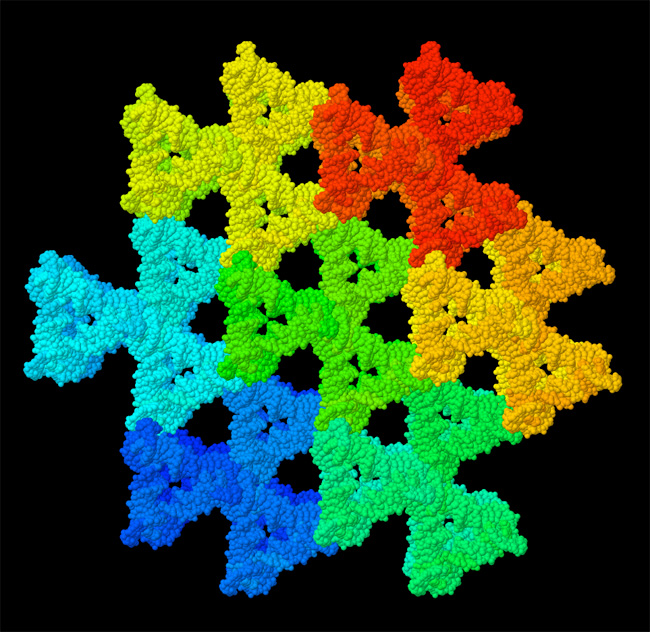
Biomolecular structures have also opened up a new discipline of biomolecular engineering and bionanotechnology. With understanding comes control, and researchers are currently modifying existing biomolecules for new functions, or even designing entirely new biomolecules. For instance, scientists are designing and building nanoscale structures with DNA (Figure 3). One of the great challenges in biomolecular engineering is the prediction of protein structure from the amino acid sequence of the chain. Great steps have been made toward solving this challenging problem, building largely on the huge database of available protein structures, but a comprehensive solution still eludes the research community.
You Can Explore Biomolecular Structures at the RCSB Protein Data Bank
The wwPDB data centers enable anyone to explore first-hand the structure of biomolecules in the PDB archive. Much of the scientific evidence supporting the major concepts in cell and molecular biology is held in the PDB archive, and visitors can go directly to the structures and explore the atomic basis of a molecule's function. However, since the PDB is foremost a repository of scientific data, it is not laid out like a textbook, with subjects neatly ordered and examples carefully presented. At its center is the archive of atomic coordinates. These files include the atomic coordinates as well as detailed annotations describing the biology and experimental details. Users have two potential challenges: first, finding a structure file that includes the desired biomolecule, and second, displaying and exploring the structure file in a way that shows the property of interest. The RCSB (Research Collaboratory for Structural Bioinformatics) PDB site at www.pdb.org offers tools and resources to help meet these challenges (Berman et al.2000).
How Do I Find Structures in the RCSB PDB?
Many tools are available at the RCSB PDB to search a database of PDB entries. The major tool is a comprehensive search function that builds queries using a variety of properties, from names of molecules to amino acid sequences. This is complemented by other approaches to browse through structures, including sequence similarity searches, searches on keywords from the Genome Ontology project (Ashburner et al. 2000), and descriptions of domain structures. Since there are tens of thousands of structures available, finding the structure that you are interested in can be difficult. The RCSB PDB's Molecule of the Month series is designed to help with this. It highlights in detail a different molecule each month, and links to representative structures from the database.
How Do I View an Atomic Structure?
Once a suitable structure entry is found, many different molecular graphics programs may be used to display and explore the structure. Several options are available at the RCSB PDB. Jmol is currently the most popular, and was used to create the illustrations in this paper. Once the Jmol viewer is launched in your web browser, several buttons allow you to switch between the most popular representations of the biomolecule, and a scripting window lets you customize your view (Figure 4). If you want to create more advanced and elaborate illustrations, you can download the atomic coordinates to your computer and then use free programs like PyMol (DeLano 2002) or Chimera (Pettersen et al. 2004).
Understanding Scientific Data
The PDB is an exciting database to explore, but since it includes actual data from scientists, one needs to be a bit careful. PDB structure files include a few idiosyncracies that are related to the complex processes used to solve the structures. For instance, X-ray crystallographers will do almost anything to get their molecules to crystallize, so the structures may have pieces missing or sulfur atoms replaced by selenium. The file may also include only part of a whole molecule, as in the case of symmetrical molecules solved by crystallography, or many copies of the same molecule, as in the multiple models often generated in NMR spectroscopy. If you run into these challenges when you're exploring, you can consult the "Understanding PDB Data" pages at the RCSB PDB. It describes the experimental details of structure determination, how they are manifested in PDB files, and how to deal with them when exploring atomic structures.
References and Recommended Reading
Ashburner, M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25(1), 25–29 (2000).
Berman, H. M., Henrick, K. & Nakamura, H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 10(12), 980 (2003).
Berman, H. M., et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Bernstein, F. C., et al. Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 (1977).
DeLano, W. The PyMOL molecular graphics system, ., on World Wide Web (2002).
Harrington, D. J., Adachi, K. & Royer, Jr., W.E. The high resolution crystal structure of deoxyhemoglobin S. J Mol Biol. 272(3), 398–407 (1997).
Harris, L. J., et al., Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 36(7), 1581–1597 (1997).
Jmol: an open-source Java viewer for chemical structures in 3D.
Mahalingam, B., et al. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur J Biochem. 271(8), 1516-1524 (2004).
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 25(13), 1605-1612 (2004).
Zheng, J., et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 461(7260), 74-77 (2009).






























