« Prev Next »

Scientists Can Analyze Gene Function by Deleting Gene Sequences

What can mice reveal about human gene function?
On the outside, humans and mice look nothing alike. However, human and mouse chromosomes share many of the same genes. In fact, 99% of the 20,000 to 25,000 genes in humans have a similar mouse counterpart. This high degree of genetic similarity between humans and mice offers researchers a unique approach for understanding human gene functioning.
Ethical considerations prevent researchers from genetically engineering humans that lack a given gene for the sole purpose of learning how that gene functions. However, because so many mouse genes are similar to human genes, geneticists can generate knockout mice in which the mouse counterpart of a human gene of interest is deleted or disrupted. The term "knockout mouse" may at first conjure images of a mouse boxing champion or beauty queen. However, geneticists use the term to refer to a mouse that has been genetically engineered such that at least one of its genes is functionally inactivated (i.e. the inactivated gene is "knocked out"). If a mouse gene has a high degree of similarity to a human gene, researchers can predict that the two genes carry out related functions. Therefore, by generating a knockout mouse without a gene of interest, these scientists may be able to determine the functions carried out by the related human gene.
In some cases, knockout mice exhibit phenotypes that mimic symptoms associated with human disease, including cancer, diabetes, obesity, cardiovascular disease, and neurodegenerative disease. In these cases, a knockout mouse is referred to as a mouse model of that form of human disease.
Making a knockout mouse: Step by step
Step 1: Generating a targeting vector
The first step in making a knockout mouse is identifying the region of the gene that will be deleted. Because the entire mouse genome sequence is already known, it is relatively simple to look up the chromosomal location and nucleic acid sequence of the gene of interest. Once the segment of the gene that will be deleted has been mapped out, the nucleic acid sequences of the DNA segments that appear on the chromosome before and after that gene must also be identified.
Once these tasks are completed, a targeting vector specifically tailored to the gene of interest is made (Figure 1). A targeting vector is a long stretch of DNA made up of smaller pieces of DNA that have been joined together. Next, in order to make the targeting vector detectable, the scientists insert a marker gene into the middle of the vector; this marker is in some way able to "report" when it is present in a cell.
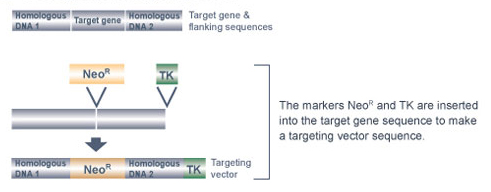
Currently, the neomycin-resistance gene, called NeoR, is a popular marker gene of choice for generating knockout mice. The antibiotic neomycin is toxic to mouse cells because they do not normally contain the NeoR gene. However, when the NeoR gene is added to mouse cells, these cells can survive in the presence of neomycin. Within the targeting vector, the NeoR gene is located between two other pieces of DNA: the "right arm" and the "left arm" of the targeting vector. The right arm of the targeting vector contains DNA with a nucleic acid sequence that matches the stretch of DNA immediately before the gene segment that will be deleted. The left arm of the targeting vector contains DNA with a nucleic acid sequence that matches the stretch of DNA immediately after the gene segment that will be deleted. The right and left arms of the targeting vector facilitate homologous recombination between the targeting vector and the target gene, thereby enabling the NeoR gene to replace the target gene segment.
The targeting vector also contains one additional piece of DNA, called a negative selection marker gene. This gene is located at the right end of the targeting vector, after the right arm. The thymidine kinase (TK) gene from the herpes simplex virus is the most commonly used negative selection marker gene. Normally, mouse cells can grow in the presence of the antiviral drug ganciclovir. The TK gene is considered a "cell suicide gene," however, as cells containing the TK gene convert ganciclovir into a lethal toxin.
Why is it necessary to include a cell suicide gene as part of the targeting vector? The reason is purely a matter of identification - specifically, the TK gene helps researchers locate cells that have correctly replaced the targeted gene segment with the NeoR gene. Often, mouse cells randomly insert the targeting vector in the wrong chromosomal location. If random insertion occurs, both the NeoR gene and the TK gene are inserted into the genome. As a result, the cells are resistant to neomycin, but they die in the presence of ganciclovir. In comparison, when the targeted gene segment is correctly replaced, the TK gene is not inserted into the chromosome along with the NeoR gene, so the resultant cells are resistant to both neomycin and ganciclovir. Therefore, the presence of the TK gene in the targeting vector allows researchers to efficiently screen for mouse cells that have correctly replaced the targeted gene segment by growing these cells in the presence of both neomycin and ganciclovir.
Step 2: Inserting the target sequence and selecting cells with the insertion
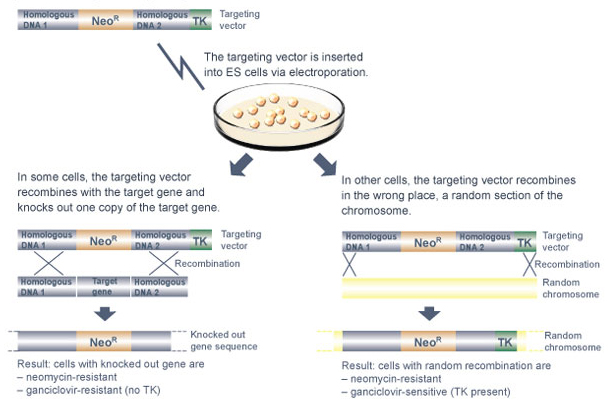
After the targeting vector is made, it is used to knock out one copy of the target gene in mouse embryonic stem (ES) cells (Figure 2). But why must mouse ES cells be used? Why can't the targeting vector be introduced into any type of mouse cell?
It is certainly possible to use the targeting vector to knock out one of the two copies of the target gene in a standard somatic cell. However, unless that cell is an ES cell, the knockout mutation cannot be incorporated into a growing embryo. Therefore, it would not be possible to study the effects of the knockout mutation in a developing mouse.
What makes ES cells so special? Primarily, it is their ability to become any one of the different adult cell types. When injected into a mouse embryo, the ES cells themselves are capable of maturing into some of the tissues of the developing mouse.
But how, exactly, are targeting vectors delivered into ES cells? Most often, a technique called electroporation is used. When ES cells are electroporated, a brief pulse of an electrical field is applied to the outside of the cells, creating a momentary increase in plasma membrane permeability and allowing the uptake of foreign DNA into the ES cells.
After the ES cells have been electroporated, they are grown in the presence of neomycin to select for those particular cells that have taken up the targeting vector. Next, the neomycin-resistant cells are grown in the presence of ganciclovir to select for those that have inserted the targeting vector at the correct location within the mouse genome.
Step 3: Identifying ES cells with the correct gene knocked out
Step 4: Injecting heterozygous knockout ES cells into a developing embryo and transferring the embryo into a mouse

Still, only some of the cells that make up the chimeric mouse carry the knockout mutation, and those cells are heterozygous for the mutation. In order to learn about the function of the target gene, a mouse that is homozygous for the knockout mutation must be studied.
Step 5: Mating chimeric mice to yield homozygous knockout mice
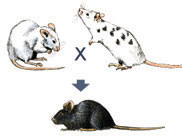
These chimeric mice can be identified by crossing them with normal white mice. If black offspring are produced from such a cross, a chimeric mouse is capable of passing the knockout mutation on to its offspring. When this is the case, 50% of the black offspring are heterozygous for the knockout mutation in all of their cells (Figure 6).
Standard molecular biology techniques can be used to determine which of the black offspring are heterozygous for the knockout mutation. Then, to produce a homozygous knockout mouse, a heterozygous knockout male is mated to a heterozygous knockout female (Figure 7). Twenty-five percent of the resulting offspring will be homozygous knockout mice, which can, again, be readily identified using standard molecular biology techniques.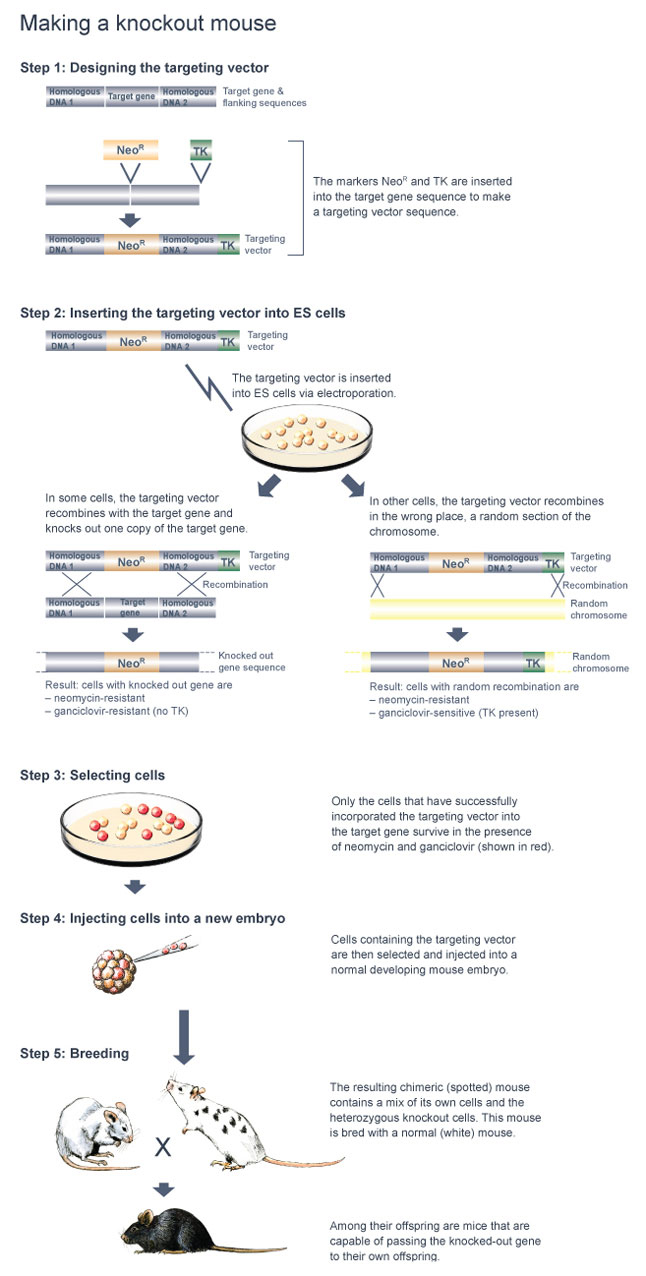
Step 6: Phenotypic characterization of homozygous knockout mice
After the homozygous knockout mice have been created, researches must then characterize the phenotypes associated with the loss of the target gene. In theory, every measurable phenotype must be examined in order to determine every possible function of the knocked-out gene. The measurement of a given phenotype in a knockout mouse would then be compared to measurements of the same phenotype in a wild-type mouse (a mouse that has not been genetically engineered for specific traits) in order to identify the functions that are altered in the knockout mouse.
Phenotypes such as size, weight, metabolism, behavior, bone development, neurological function, reproduction, and aging can be easily measured. If the knocked-out gene is required for development, however, it may not be possible to produce homozygous knockout mice. In this case, researchers may study heterozygous knockout mice, or they may instead turn to other types of knockout mice, such as conditional knockout mice (in which the target gene is inactivated in response to a specific stimulus) or tissue-specific knockout mice (in which the target gene is inactivated in only one or several tissues).
Large-scale mouse knockout projects
In September 2006, the U.S. National Institutes of Health (NIH) initiated a five-year, $52 million project called the Knockout Mouse Project (KOMP). Together with the Texas A&M Institute of Genomic Medicine (TIGM), the North American Conditional Mouse Mutagenesis Project (NorCOMM) in Canada, and the European Conditional Mouse Mutagenesis Program (EUCOMM), KOMP set the goal of knocking out every one of the 20,000 mouse protein-encoding genes within five years.
Similar to the mouse knockout consortia mentioned above, a number of labs have also collaborated to establish standardized methods for the phenotypic characterization of knockout mice. The European Union Mouse Research for Public Health and Industrial Applications (EUMORPHIA) group developed the first set of standard phenotyping protocols, which was validated among several different labs. The European Mouse Phenotypic Resource for Standardized Screens (EMPReSS) then established the primary phenotypic screen used by the European knockout mouse labs, one that comprises a subset of the standard protocols of EUMORPHIA.
Future directions
Further Exploration
Key Questions
eBooks
This page appears in the following eBook





















