« Prev Next »
Kinetoplastids are flagellated protozoans, which are unicellular eukaryotic organisms. They include free-living microorganisms, as well as parasites of diverse invertebrate, vertebrate, and plant species. Some kinetoplastids are responsible for serious human diseases, such as Chagas disease and sleeping sickness (caused by Trypanosoma cruzi and Trypanosoma brucei, respectively), and the various forms of cutaneous and visceral leishmaniasis (caused by Leishmania spp.). All kinetoplastids share a unique mitochondrial DNA structure called kinetoplast DNA, or kDNA. Unlike any other type of DNA in nature, kDNA is organized in a unique giant network of interlocked rings.
The network of rings in kDNA forms a beautiful structure. Observed under the electron microscope, it resembles the chainmail that medieval knights wore under their plate armor for protection. In their research on kDNA, scientists eager to understand the function of this amazing structure made a second unexpected discovery: a novel, complex mechanism for RNA processing of mitochondrial transcripts, now known as RNA editing. Investigators from different disciplines have been working hard over the past decades to gather clues to explain the unusual properties of kDNA. Today there are still many questions left to answer.
The Kinetoplast: An Unusual Structure for Mitochondrial DNA
The first tools developed for the study of cells and tissues were optical microscopes. These microscopes use visible light and one or more lenses to magnify small objects. Since their invention about 400 years ago, scientists have constructed different models and types of light microscopes. In the early twentieth century, with the aid of photographic cameras, microscopists obtained the first micrographs of cell structures. Under ideal conditions, an optical microscope enables us to observe objects as small as 0.2 micrometer (mm; 1 micrometer is equal to one-millionth of a meter).
It was under the light microscope that scientists first recognized the kinetoplast as a structure that stains with basic dyes inside the single mitochondrion of certain flagellates. Trager described the structure as "a small spherical or rod-shaped structure lying just posterior to the basal body of the flagellum" (Figure 1; Trager 1965). At the time, scientists knew that the kinetoplast was Feulgen positive (the Feulgen stain is a staining technique that specifically identifies DNA). In addition, they knew it incorporated tritiated thymidine, a radioactive form of thymidine, which is a chemical compound found in DNA (Steinert et al. 1958). Both of these observations strongly suggested that this structure contained mitochondrial DNA. Scientists used radioactive incorporation of thymidine and autoradiography to show that the kinetoplast is a self-replicating organelle and that its division precedes that of the nucleus. Moreover, the kinetoplast reacted to specific chemical reactions for mitochondria.
Although scientists were biochemically characterizing the kinetoplast, the beautiful chainmail structure of the kDNA would remain hidden until the 1970s, when several investigators discovered its two-dimensional fishnet-like network (Figure 2). When condensed, kDNA has a disc-shaped planar structure. The discovery of its two-dimensional structure was possible thanks to the invention of the electron microscope. This revolutionary visual aid enabled researchers to view structures a thousand times smaller than those observed with the light microscope (electron microscopes allow us to observe objects as small as 0.0002 mm). When scientists isolated the kinetoplast using density gradients and centrifugation, collected the DNA content on grids, and studied it under the electron microscope, they discovered that the kDNA contained two different kinds of DNA (Laurent & Steinert 1970; Simpson & Da Silva 1971). What were these two types of structures?
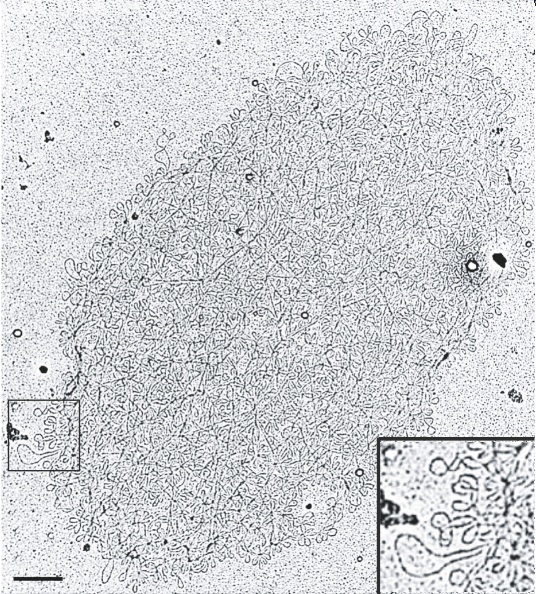
Today researchers know that the kDNA network is about 10 mm by 15 mm and is composed of two types of DNA rings, consisting of 5,000–10,000 minicircles and 25–50 maxicircles. Depending on the species, the minicircles range from 0.5 to 10 kilobases (kb) in size, whereas maxicircles are approximately 20–40 kb. (The kilobase is a unit of measure used in molecular biology that is equal to 1,000 nucleotide base pairs of DNA). All of the DNA minicircles and maxicircles are interlocked in a huge DNA catenane, which gives kDNA its characteristic chainmail structure. How did scientists discover this unique structure? They received help from some unusual allies — parasites.
Trypanosomatids as Laboratory Models
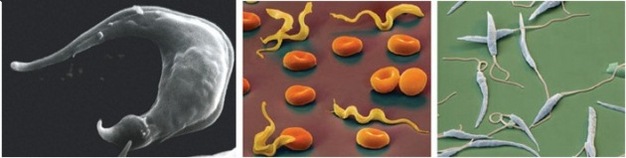
When we think of parasites, especially human parasites, often what first comes to mind are the diseases they cause. From the cell biology perspective, however, parasites are extraordinarily useful experimental models from which scientists have been able to discover exciting new information about general biology. As an example, most of what we know about the structure and replication of kDNA comes from studies of trypanosomatids. Scientists have studied kDNA in Crithidia fasciculata (in insect parasite) and Leishmania tarentolae (a lizard parasite), which are nonpathogenic to humans and easy to cultivate in the laboratory. In addition, scientists have performed many studies in trypanosomatids that cause human diseases, including T. brucei, T. cruzi, and Leishmania spp. (Figure 3).
In electron micrographs of the intact isolated kDNA network, scientists had difficultly discerning how the chained together (catenated), fishnet-like network was organized. To study the kDNA structure, scientists used topoisomerases (enzymes that make specific cuts in the DNA) to decatenate the rings. This treatment, along with photography in platinum and palladium, finally made it possible to visualize both the minicircles and the maxicircles. One of the most studied networks was that of Crithidia fasciculata. Its kDNA network contains approximately 5,000 minicircles of 2.5 kb each and about 25 maxicircles of 37 kb each. Scientists determined the size of both the minicircles and the maxicircles through careful DNA treatments with restriction enzymes and DNA sequencing. Sequencing revealed that the minicircles are heterogeneous in sequence.
Within the network, each minicircle is connected to three neighboring minicircles through a single interlock. This chainmail-like network is quite flexible and can be organized in a planar sheet. In contrast, the maxicircle catenanes are organized as rosettes with a dense central core and protruding loops. Both catenanes are extensively linked with one another to form a complicated kDNA network or "network within a network" (Shapiro 1993).
Scientists hypothesized that proteins were involved in kDNA condensation within the mitochondrial matrix (in the same way that basic proteins known as histones help compact and organize nuclear DNA). To prove this idea, researchers designed a very interesting experiment. They used formaldehyde to cross-link proteins to kDNA in a reversible fashion. Commonly used to preserve or fix tissues for staining, formaldehyde has the ability to produce links between amino groups in proteins and other nearby nitrogen groups in molecules like DNA or other proteins. These links can be easily broken with heat. After linking proteins to the kDNA in whole C. fasciculata cells, the investigators purified the network and observed that it remained cross-linked and condensed. The network expanded only if treated with proteinases (enzymes that degrade proteins) or if the cross-linking was reversed by heating the network to 65°C. Using these experiments, scientists demonstrated that proteins were present in the condensed network. Moreover, they isolated five small proteins that were released from the kDNA following the heat treatment. By using biochemical and sequencing analyses, they learned that two of the proteins were basic proteins (Xu & Ray 1993). These proteins organize the kDNA into its characteristic disc-like shape
The Moving Kinetoplast
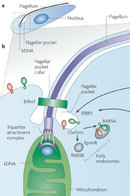
Changes in the position of the kDNA are used to identify the several differentiated forms of the parasite (Figure 5). Scientists were curious why the kinetoplast remained near the basal body. Was there a physical connection between the basal body and the kDNA?
To answer this question, researchers pioneered a technique, called quick-freeze deep-etch electron microscopy, that enabled them to take detailed pictures of short-lived events inside living cells. Cells from cultured T. cruzi and Herpetomonas megaseliae (an insect parasite) were quickly frozen in a liquid helium-cooled machine and then fractured to view their internal structures. The technique yielded three-dimensional images for analysis of the kinetoplast architecture. The investigators observed that "filamentous structures connected the kinetoplast to the basal body." Other studies confirmed "a physical linkage between the kinetoplast and the basal body" (Figure 6; Souto-Padron et al. 1984; Gull 1999).
Mitochondrial DNA
Kinetoplast DNA represents approximately 30 percent of the total cell DNA. It differs from nuclear DNA in several properties, including its buoyant density. Since the size of the maxicircle is comparable in size to the mitochondrial DNA of other eukaryotic cells, scientists assumed that mitochondrial information was contained in these rings. Using DNA sequencing to analyze the kDNA, they learned, as expected, that maxicircles encode typical mitochondrial proteins, such as cytochrome b, cytochrome oxidase subunits, NADH dehydrogenase subunits, ATPase subunit 6, and ribosomal RNA.
A new question immediately arose: If the mitochondrial DNA was encoded in the maxicircles, what was the function of the minicircles? Did they have any function at all? Any transcription and translation of the sequences contained in these small rings would produce a very small protein. When scientists began studying the transcription of the mitochondrial genes, they found unexpected clues to answer these question.
RNA Editing
Soon, other kinetoplastid mitochondrial transcripts were shown to correct frameshifts in a similar fashion with uridine insertions and sometimes deletions. In addition, some mRNA sequences were altered through uridine insertions that added AUG initiation codons. These discoveries provided further support for RNA editing. Incredibly, the discovery of this new cellular mechanism did not catch the attention of biologists around the globe until 1988, when Kenneth Stuart and his collaborators showed that over 50 percent of the T. brucei cytochrome oxidase subunit III (coxIII) transcript underwent RNA editing. How did these researchers discover the massive RNA editing of coxIII?
The first studies, which used both analysis of the nucleotide sequence and cross-hybridization studies, were not successful in detecting the coxIII gene. However, by RNA and cDNA sequencing, scientists were able to identify the mRNA transcript in the mitochondrion of T. brucei. When scientists compared the nucleotide and predicted amino acid sequences they found them to be very similar to other coxIII genes from related species. Knowing of Benne et al.'s experiments, Stuart and his group decided to examine the mitochondrial DNA. They suspected that the transcript sequence was created by RNA editing. As expected, they found a genomic sequence corresponding to the coxIII transcript, but lacking several thymidines (these thymidines are substituted for uridines during mRNA transcription). In the end they found that, of the 1,000 nucleotides of the mature coxIII mRNA, over 50 percent of the sequence consisted of uridine residues that had been added or deleted. In over 150 separate sites, uridine residues were added, while at least 19 were deleted from 9 separate sites. This massive editing explained why scientists had so much trouble detecting the the coxIII gene in T. brucei's genome (Feagin et al. 1988). Finally, RNA editing caught the attention of other researchers.
However, the absence of a molecule that could serve as a template to guide the uridine insertions and deletions provoked skepticism of this novel cellular process. The discovery, made by Larry Simpson and his colleagues, of short (40–80 bases) mitochondrial transcripts that are complementary to edited mRNA sequences represented the first breakthrough in understanding the RNA editing mechanism. These short transcripts, which they termed "guide RNA" (gRNA), presumably directed the editing process. Simpson's group discovered that the gRNA molecules were mainly encoded by minicircles, although some gRNAs can also be encoded by maxicircles. At last, scientists had discovered a function of the minicircles in the kDNA network.
Guide RNA contains a segment that is complementary to a region of the edited version of a transcript. This complementary segment is located at the 5' end of the gRNA and is known as the anchor sequence, which provides a template for the insertion and deletion of uridines. In addition, most gRNA has an poly-U tail (5–25 nucleotides) on its 3' end, which provides a source of uridines that can be added to mRNA, or a site to accept uridines following their deletion from the mRNA (Blum et al. 1990). To date, the only known function of the minicircles is to encode gRNAs. Organisms that extensively edit their mRNA transcripts, such as T. brucei, possess as many as 200 different minicircles with a variety ofsequences in their kDNA network.
The identification of gRNA as the template for RNA editing quickly led scientists to formulate two alternative models to explain how the accurate insertion and/or deletion of nucleotides is accomplished. One model was the cleavage-ligation (cut-and-paste) pathway; the second was the transesterification pathway (in which organic groups are exchanged). Scientists developed in vitro systems to carry out uridine deletion and insertion under controlled conditions in the laboratory to test both models, and the results favored the cleavage-ligation model.
Understanding RNA Processing
Kinetoplastids have helped scientists understand RNA processing events, such as RNA editing. To date, gene expression resulting from RNA editing has been described in organisms as diverse as unicellular protozoans, fungi, nematodes, viruses, and humans. Our modern concept of RNA editing involves not only the insertion and deletion of nucleotides, but also the conversion of one base to another. In any case, these processes seem to occur via unrelated mechanisms. Thanks to the kinetoplastids, we now have a clearer picture of RNA processing events. However, there remain many questions related to the different RNA editing mechanisms and the evolutionary implications of these processes.
Summary
The kDNA structure itself is still a subject of intense research and debate. Electron microscopy studies that began in the 1970s revealed that some kinetoplastids, including both free-living and parasitic species, have somewhat different kDNA structures. In some species, the kDNA does not appear condensed in one region, but scattered throughout the mitochondrial matrix. And quite unexpectedly, scientists working with Bodo caudatus (a nonparasitic, free-living kinetoplastid) found that its kDNA contains minicircles that are not chained together (Hadjuk et al. 1986). To date, there is general agreement among scientists that kDNA can exist in a vast diversity of structures, and now biologists are trying to understand the biological significance of the kDNA network, different kDNA structures, and the role of kDNA in parasitism.
References and Recommended Reading
Benne, R. et al. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826 (1986).
Blum, B., Bakalara, N. & Simpson, L. A model for RNA editing in kinetoplastid mitochondria: RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60, 189–198 (1990).
Feagin, J. E., Abraham, J. M. & Stuart, K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53, 413–422 (1988).
Gull, K. The cytoskeleton of trypanosomatid parasites. Annual Review of Microbiology 53, 629–655 (1999).
Hadjuk, S. L., Siqueira, A. M. & Vickerman, A. K. Kinetoplast DNA of Bodo caudatus: A noncatenated structure. Molecular and Cellular Biology 6, 4372–4378 (1986).
Laurent, M. & Steinert, M. Electron microscopy of kinetoplastic DNA from Trypanosoma mega. PNAS 66, 419–424 (1970).
Shapiro, T. A. Kinetoplast DNA maxicircles: Networks within networks. PNAS 90, 7809–7813 (1993).
Simpson, L. & Da Silva, A. Isolation and characterization of kinetoplast DNA from Leishmania tarentolae. Journal of Molecular Biology 56, 443–473 (1971).
Souto-Padron, T., de Souza, W. & Heuser, J. E. Quick-freeze, deep-etch rotary replication of Trypanosoma cruzi and Herpetomonas megaseliae. Journal of Cell Science 69, 167–178 (1984).
Steinert, G., Firket, H. & Steinert, M. Synthesis of deoxyribonucleic acid in the parabasal body of Trypanosoma mega. Experimental Cell Research 15, 632–635 (1958).
Trager, W. The kinetoplast and differentiation in certain parasitic protozoa. American Naturalist 99, 255–266 (1965).
Xu, C. & Ray, D. S. Isolation of proteins associated with the kinetoplast-DNA networks in vivo. PNAS 90, 1786–1789 (1993).



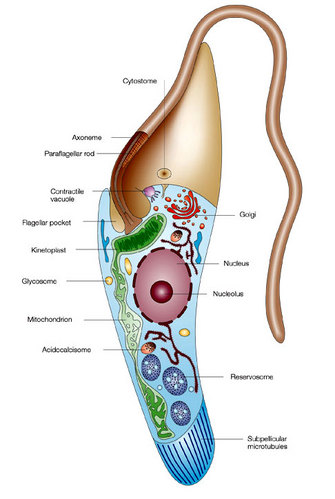
 Figure 1: Schematic representation of trypanosomatids.
Figure 1: Schematic representation of trypanosomatids.