« Prev Next »
Replicating DNA is fragile, and can break during the duplication process. In fact, broken chromosomes are often the source of DNA rearrangements and can change the genetic program of a cell. These changes can trigger a growth advantage in a single cell in your body, and when that cell continues to divide, tumors arise. Fortunately, our cells have defense mechanisms to shield us from these damaging events.
In the eukaryotic cell cycle, chromosome duplication occurs during "S phase" (the phase of DNA synthesis) and chromosome segregation occurs during "M phase" (the mitosis phase). During S phase, any problems with DNA replication trigger a ‘'checkpoint" — a cascade of signaling events that puts the phase on hold until the problem is resolved. The S phase checkpoint operates like a surveillance camera; we will explore how this camera works on the molecular level. The last 60 years of research in bacterial species (specifically, Escherichia coli) and fungal species (specifically, Saccharomyces cerevisiae), have continually demonstrated that several major processes during DNA replication are evolutionarily conserved from bacteria to higher eukaryotes.
A Review of the Proteins and Process of DNA Replication
Before delving into the intricacies of checkpoints, we must remind ourselves of the key molecules and processes of DNA replication. What happens to DNA when it is duplicated?
The Replication Fork and DNA Strand Orientation
Recall that chromosomes are made of double-stranded (ds) DNA. How does the cell duplicate two strands of identical DNA copies simultaneously? The goal of replication is to produce a second and identical double strand. Because each of the two strands in the dsDNA molecule serves as a template for a new DNA strand, the first step in DNA replication is to separate the dsDNA. This is accomplished by a DNA helicase. Once the DNA template is single-stranded (ss), a DNA polymerase reads the template and incorporates the correct nucleoside-triphosphate in the opposite position (Figure 1). Because of the characteristic y-shape of the replicating DNA, it is often referred to as a "replication fork." Particularly important are two aspects of the replication fork: 1) the 5' to 3' polarity of the newly synthesized DNA and 2) the sequence of base pairs (color-coded in Figure 1). The DNA code in each of the strands is the same, but inverted, so that the sequence is identical when read in the 5' to 3' direction. This is the direction in which all DNA is polymerized, and also the direction in which a DNA sequence is read when written out, by convention.
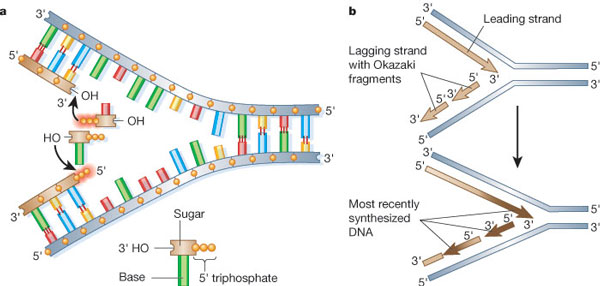
The Leading and Lagging Strands
The DNA strand that is synthesized in the 5' to 3' direction is called the leading strand. The opposite strand is the lagging stand, and although it is also synthesized in the 5' to 3' direction, it is assembled differently. As a rule, none of the known DNA polymerases adds a nucleoside triphosphate onto a free 5' end. This brings us to the first rule of DNA replication: DNA synthesis only occurs in one direction, from the 5' to the 3' end.
Applying this rule helps us understand why the lagging strand is generated from a series of smaller fragments (Figure 1b). These fragments are known as Okazaki fragments, after Reiji and Tsuneko Okazaki, who first discovered them in 1968. Each time the DNA fork opens, the leading strand can be elongated, and a new Okazaki fragment is added to the lagging strand. All Okazaki fragments are subsequently joined together by DNA ligase to form a long continuous DNA strand (Anderson & DePamphilis 1979; Alberts 2003). In this regard, eukaryotic DNA replication follows the same principles as prokaryotic DNA replication.
The Key Proteins at the Replication Fork: Polymerases, Primases, and Helicases
Amongst the array of proteins at the replication fork, DNA polymerases are central to the process of replication. These important enzymes can only add new nucleoside triphosphates onto an existing piece of DNA or RNA; they cannot synthesize DNA de novo (from scratch), for a given template. Another class of proteins fills this functional gap. Unlike DNA polymerases, RNA polymerases can synthesize RNA de novo, as long as a DNA template is available. This particular feature of de novo synthesis is similar to what happens during mRNA transcription.
Eukaryotic cells possess an enzyme complex that has RNA polymerase activity, but works in DNA replication. This unique enzyme complex is called DNA primase. Interestingly, this primase generates small 10-nucleotide-long RNA primers from a DNA template (the red portion of the Okazaki fragment in Figure 2). The RNA primers produced are later replaced by DNA, so that the newly-synthesized lagging strand is not a mixture of DNA and RNA, but consists exclusively of DNA. The chemical properties of DNA and RNA are quite different, and DNA is the preferred storage material for the genetic information of all cellular organisms, so this reinstallment of a continuous DNA strand is very important.
In prokaryotic cells, DNA primase is its own entity and works in a complex with the DNA helicase (Figure 2) (Alberts 2003; Langston & O'Donnell 2006). However, in eukaryotic cells DNA primase is associated with another polymerase, DNA polymerase-α | | | pol-α | | |, which initiates the leading strand and all Okazaki fragments (Pizzagalli, A. et al. 1988; Hubscher, Maga, & Spadari 2002). At present, we have no evidence that DNA primase binds to the DNA helicase in eukaryotic cells. But it is likely that some connector protein coordinates DNA unwinding and DNA synthesis initiation in eukaryotic cells.
After strand initiation, other DNA polymerases continue DNA elongation. In eukaryotic cells, these polymerases cooperate with a sliding clamp called proliferating cell nuclear antigen (PCNA). The regulation of PCNA is highly complex and important for DNA replication and repair (Moldovan, Pfander, & Jentsch 2007). There may be additional, yet undiscovered, parallel (or identical) mechanisms or proteins that coordinate DNA unwinding and DNA elongation. Observations in simpler model organisms strongly hint that eukaryotes too have a connecting mechanism that coordinates DNA helicase, and a DNA polymerase-a/DNA primase (pol-a/primase) complex.
Possible Connecting Links Between DNA Unwinding and DNA Synthesis
How would you identify the protein that serves as a connector between DNA helicase and pol-a/primase? A simple yet often effective approach is to find proteins that directly bind to both enzymes. However, that requires us to understand the molecular architecture of DNA helicase.
In eukaryotes, the DNA helicase is comprised of a structural core and two regulatory subunits. The core, which contains the ATP hydrolysis activity, is a hexameric complex formed of the minichromosome maintenance proteins 2-7, called Mcm2-7 (Bochman & Schwacha 2008; Bochman & Schwacha 2009; Schwacha & Bell 2001). Mcm2-7 encircles dsDNA (Remus et al. 2009), but remains inactive until two additional regulatory subunits assemble onto it. Those factors are cell division cycle protein 45 (Cdc45) and GINS (Go, Ichi, Ni, and San; Japanese for "five, one, two, and three," which refers to the annotation of the genes that encode the complex). Scientists call this resulting functional DNA helicase a CMG complex (formed by Cdc45, Mcm2-7, GINS) (Moyer, Lewis, & Botchan 2006). In principle, any of these assembled components could be linked to pol-a/primase by a hypothetical connector protein. Scientists have actually identified two candidate connector proteins that directly bind to both helicase and primase: 1) Mcm10 (another Mcm protein that, despite its name, has no functional resemblance to any of the Mcm2-7 proteins) (Solomon et al. 1992.; Merchant et al. 1997) and 2) chromosome transmission fidelity protein 4 (Ctf4) (Kouprina et al. 1992). Specifically, both of these proteins interact with pol-a/primase (Fien et al. 2004; Ricke & Bielinsky 2004; Warren et al. 2009; Miles & Formosa 1992) and CMG complex subunits (Merchant et al. 1997; Gambus et al. 2009). In budding yeast, Mcm10 is essential for replication to occur. However, in these same cells DNA replication can function normally without Ctf4, which means that Ctf4 is not absolutely required (Kouprina et al. 1992). What about higher eukaryotes? Other experiments in human cells have shown that both proteins seem to be necessary, and work together during replication (Zhu, et al. 2007). Scientists are still actively investigating these complex mechanisms.
Coordination of DNA Unwinding and Replication is Essential
Why is coordination between DNA unwinding and synthesis important? What would happen if you lose this coordination? Because pol-a/primase always requires CMG function to create the ssDNA template, it could never surpass the DNA helicase (Figure 2b). Without a connecting link, the CMG complex could just "run off" and leave pol-a/primase behind. This would create long regions of vulnerable ssDNA. Therefore, the second rule in DNA replication is that DNA unwinding and DNA synthesis have to be coordinated.
Triggering a Checkpoint
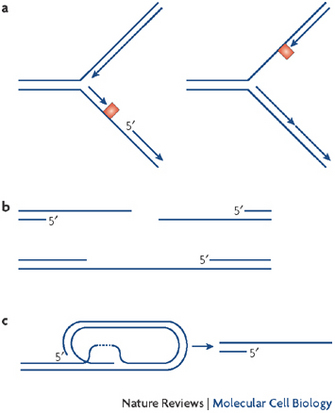
What is the mechanism of a red flag, or danger signal that activates a checkpoint? How does it alert the cell? Scientists who have asked this question don't know the entire answer, but they have learned that RPA-coated ssDNA attracts a specific protein with a complicated name: the ataxia telangiectasia mutated and Rad3 related kinase, also known as ATR (Cimprich & Cortez 2008). ATR associates with RPA and activates its intrinsic kinase activity. This starts a that temporarily halts S phase progression. Therefore, ATR is also known as the S phase "checkpoint kinase."
ATR kinase acts in several ways to keep the replication process intact. There is evidence that ATR also stabilizes replication forks that contain ssDNA (Katou et al. 2003). How this happens remains largely unclear, but recent evidence suggests that ATR may affect the Mcm2-7 proteins, the inner core of the CMG helicase mentioned above (Cortez, Glick, & Elledge 2004; Yoo et al. 2004). One hypothesis is that phosphorylation of one or several of the Mcm2-7 subunits prevents the CMG complex from unwinding more and more DNA. This action effectively stops the process so that it can be repaired before proceeding. Currently, many researchers are trying to better understand the mechanisms of crosstalk between ATR and the replication machinery (Forsburg 2008; Bailis et al. 2008).
Other Roles for ATR
Stalled Forks
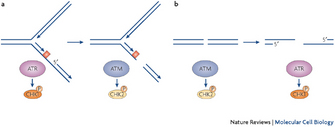
Scientists use the term "stalled forks" for areas of replication forks where DNA polymerization is halted. Stalled forks activate ATR, which in turn phospohorylates its downstream target, the checkpoint kinase 1 (Chk1) (Figure 4) (Cimprich & Cortez 2008). Little is known about the phosphorylation targets that lie further downstream of Chk1, but when scientists observe Chk1 phosphorylation in cells, they conclude that cells are actively trying to protect replication forks with DNA lesions.
Double-Stranded Breaks (DSBs)
What happens when ATR function goes awry? Normally, once DNA polymerization resumes and ssDNA is converted into dsDNA, ATR is inactivated and cells are released from the checkpoint. However, if the ATR signaling pathway is defective, due to a mutation in ATR or Chk1 (Menoyo et al. 2001), then ssDNA is converted into a double-strand break (DSB), a complete cleavage of both DNA strands (Figure 4, right).
A DSB is a catastrophic event because it ruins the replication fork. Under these circumstances, cells activate the ATM kinase (Figure 4, on the right). As mentioned above, ATM and ATR are related to each other as they share some amino acid sequences (Shiloh 2003), but ATM has a different function: it works exclusively to repair DSBs (Cimprich & Cortez 2008). It does so by phosphorylating checkpoint kinase 2 (Chk2), a protein that triggers a cascade of phosphorylation events that ultimately result in the repair of the DSB. Only if the DSB is successfully repaired can DNA replication resume.
Interestingly, when Chk2 triggers events that ultimately repair a DSB, another event also takes place. This event is the phosphorylation of the well-known p53 (Caspari 2000). This observation is a clue that repairing DSBs may have something to do with preventing the formation of tumors.
DSBs and DNA Rearrangements in Cancer Cells
Together with a variety of other molecules, ATR and ATM kinases are key factors for the surveillance of DNA replication, and prevent chromosome breakage in dividing cells. However, during repair processes, chromosome fragments can be improperly joined together. Indeed, some scientists consider that such mistakes enable some degree of genetic evolution by creating new and different genetic sequences. Nevertheless, if even a single cell in our body makes a mistake and fuses DNA fragments to each other that are not supposed to be joined, the rearrangement can be sufficient to deregulate normal cell division. If multiple changes of this type accumulate, then this single cell can eventually turn into a tumor.
Given this understanding, would it be true that people who carry a mutation in the ATM, ATR, CHK1, or CHK2 genes have a higher risk of developing cancer? Yes. In these affected individuals, the cellular surveillance system described above is defective and no longer provides full protection from random events that affect DNA replication. For example, the name of the ATM protein derives from the affliction that results from a mutated ATM protein: ataxia telangiectasia. In this disease, patients suffer from motor and neurological problems, and they also have what is known as a genome instability syndrome that genetically predisposes them to developing cancer (Shiloh 2003). In addition, when scientists examine cells directly, the experimental inhibition of ATM, ATR, Chk1, Chk2, or the connector protein Mcm10 causes a very dramatic increase of DSBs (Paulsen et al. 2009; Chattopadhyay & Bielinsky 2007). With these observations, it may be possible to create new ideas for novel diagnostics and therapies for cancer that specifically track these potent molecules.
Summary
The process of DNA replication is highly conserved throughout evolution. Investigating the replication machinery in simple organisms has helped tremendously to understand how the process works in human cells. Major replication features in simpler organisms extend uniformly to eukaryotic organisms, and replication follows fundamental rules. During replication, complex interactions between signaling and repair proteins act to keep the process from going awry, despite random events that can cause interruption and failures. Discovering the exact repair mechanisms that help keep DNA intact during replication may help us understand the mechanisms of tumor growth, as well as develop strategies to detect or treat cancer.
References and Recommended Reading
Alberts, B. DNA replication and recombination. Nature 421 431–435 (2003). doi:10.1038/nature01407.
Anderson, S. & DePamphilis, M. L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. The Journal of Biological Chemistry 254 11495–11504 (1979).
Bailis, J.M. et al. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote S-phase genome stability. Molecular and Cellular Biology 28 1724–1738 (2008) doi:10.1128/MCB.01717-07.
Bochman, M.L. & Schwacha, A. 2008. The Mcm2-7 complex has in vitro helicase activity. Molecular Cell 31 287–293. doi:10.1016/j.molcel.2008.05.020.
Bochman, M.L. & Schwacha, A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73 652–683. doi:10.1128/MMBR.00019-09.
Caspari, T. How to activate p53. Current Biology 10 R315–317 (2000) doi:10.1016/S0960-9822(00)00439-5.
Chattopadhyay, S. & Bielinsky, A. K. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage. Molecular Biology of the Cell 18 4085–4095 (2007) doi: 10.1091/mbc.E06-12-1148.
Cimprich, K.A. & Cortez, D. 2008. ATR: an essential regulator of genome integrity. Nature Reviews 9 616–627 (2007) doi:10.1038/nrm2450.
Cortez, D., Glick, G., & Elledge, S. J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proceedings of the National Academy of Sciences of the United States of America 101 10078–10083 (2004) doi: 10.1073/pnas.0403410101.
Fien, K. et al. 2004. Primer utilization by DNA polymerase alpha-primase is influenced by its interaction with Mcm10p. The Journal of Biological Chemistry 279 16144–16153. doi: 10.1074/jbc.M512997200.
Forsburg, S.L. The MCM helicase: linking checkpoints to the replication fork. Biochemical Society Transactions 36 114–119 (2008).
Gambus, A. et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. The EMBO Journal 28 2992–3004 (2009) doi:10.1038/emboj.2009.226.
Hubscher, U., Maga, G., & Spadari, S. Eukaryotic DNA polymerases. Annual Review of Biochemistry 71 133–163 (2002) doi:10.1146/annurev.biochem.71.090501.150041.
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 1078–1083 (2003) doi:10.1038/nature01900.
Kouprina, N. et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Molecular and Cellular Biology 12 5736–5747 (1992).
Langston, L. D. & O'Donnell, M. DNA replication: keep moving and don't mind the gap. Molecular Cell 23 155–160 (2006) doi:10.1016/j.molcel.2006.05.034.
Menoyo, A. et al. Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Research 61 7727–7730 (2001).
Merchant, A.M. et al. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Molecular and Cellular Biology 17 3261–3271 (1997).
Miles, J. & Formosa, T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Molecular and Cellular Biology 12 5724–5735 (1992).
Moldovan, G.L., Pfander, B., & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129 665–679 (2007) doi:10.1016/j.cell.2007.05.003.
Moyer, S.E., Lewis, P.W. & Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences of the United States of America 103 10236–10241 (2006) doi: 10.1073/pnas.0602400103.
Paulsen, R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular Cell 35 228–239 (2009) doi:10.1016/j.molcel.2009.06.021.
Pizzagalli, A. et al. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proceedings of the National Academy of Sciences of the United States of America 85 3772–3776 (1988).
Remus, D. et al.. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139 719–730 (2009) doi:10.1016/j.cell.2009.10.015.



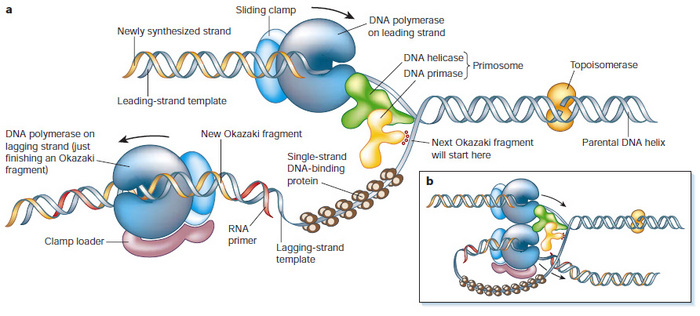
 Figure 2: Proteins at the Y-shaped DNA replication fork
Figure 2: Proteins at the Y-shaped DNA replication fork


























