« Prev Next »
Within any given population, there are multiple potential sources of phenotypic variation, and each of these sources reflects a different underlying cause. The particular source of an instance of phenotypic variation determines whether that trait has the ability to respond to natural or artificial selection (i.e., whether the trait has evolutionary potential), as well as whether the trait can respond to environmental changes. Thus, researchers are highly interested in determining the relative importance of both the genetic factors and the environmental factors that lead to particular traits. Moreover, once investigators have determined this information, they can use it to predict the evolutionary dynamics of an entire population with regard to a specific trait (Waldmann, 2001, Fisher et al., 2004, Saastamoinen, 2008).
Calculating Phenotypic Variance
As previously mentioned, all instances of phenotypic variance (VP) within a population are the result of genetic sources (VG) and/or environmental sources (VE). This relationship can be summarized as follows (Falconer & Mackay, 1996; Lynch & Walsh, 1998):
VP = VG + VE
However, in order to determine the values for both VG and VE, researchers must consider a number of additional variables, as described in the following sections.
Genetic Sources of Variation
Genetic sources of variation can themselves be divided into several subcategories, including additive variance (VA), dominance variance (VD ), and epistatic variance (VI). Together, the values for each of these subcategories yield the total amount of genetic variation (VG) responsible for a particular phenotypic trait:
VG = VA + VD + VI
Defining the Subcategories of Genetic Variance
Of course, in order to fully understand this equation, one must also understand what each subcategory of genetic variance entails. The first subcategory, additive genetic variance, refers to the deviation from the mean phenotype due to inheritance of a particular allele and this allele's relative (to the mean phenotype of the population) effect on phenotype. In contrast, the second subcategory, dominance genetic variance, involves deviation due to interactions between alternative alleles at a specific locus. For example, say that a plant produces white flowers if its genotype is A1A1 and red flowers if its genotype is A2A2. If there is no dominance deviation or interaction between these alleles, then the A1A2 genotype would lead to pink flowers due to expression of both alleles. However, if there is an interaction between the alleles such that their expression is not equal, then the phenotype of the heterozygote (A1A2) would not be an approximate midpoint; rather, it would more closely resemble one of the homozygotes (either A1A1 or A2A2). For instance, if the A2 allele was dominant to the A1 allele, then the heterozygote would produce red flowers.
Finally, like dominance variance, the third category of genetic variance—epistatic variance—involves an interaction between alleles; however, in this case, the alleles are associated with different loci. This concept can be illustrated by returning to the flower color example, but this time assuming that a second locus (denoted B) determines whether flower pigment is produced. In this case, the B1 allele is dominant, and it codes for the production of pigment; an absence of pigment means that a plant's flowers will be white. Thus, if the plant's genotype is either B1B1 or B1B2, its flower color will be determined by the genotype at the A locus. On the other hand, if the plant's genotype is B2B2, then no pigment is produced, so the flower color is always white and is not influenced by the genotype at the A locus.
Genetic Variance and Trait Heritability
Once researchers have determined the total amount of genetic variation responsible for a trait, they can use this information in calculations of the trait's heritability. Heritability is a measure of the proportion of phenotypic variance attributable to genetic variance, and it is an important predictor of the degree to which a population can respond to artificial or natural selection.
Consider, for example, a study involving the rare plant Scabiosa canescens that was conducted by researcher Patrik Waldmann in 2001. Like many rare species, this plant is often found in populations of very small size. Such populations are expected to have a low level of genetic diversity, which would limit their ability to adapt. In his study, Waldmann used genetic lines of S. canescens from both a small and a large natural population to estimate both additive and dominance genetic variation. However, contrary to expectations, Waldmann did not find a lower level of additive genetic variance in the offspring from the smaller population. This finding meant that the smaller population still had the ability to evolve, even though the population consisted of only 25 individuals.
Environmental Sources of Variation
Like their genetic counterparts, environmental sources of variation can also be placed into various subcategories; these include specific environmental variance (VEs),general environmental variance (VEg), and genotype by environment interaction (VGxE).Together, the values for each of these subcategories yield the total amount of environmental variation (VE ) responsible for a particular phenotypic trait:
VE = VEg + VGxE + VEs
Defining the Subcategories of Environmental Variance
Again, a closer understanding of each of the aforementioned subcategories is critical to an accurate interpretation of the concept of environmental variance. Here, specific environmental variance refers to the deviation from the population mean due to the environmental conditions that are uniquely experienced by each individual. Statistically speaking, this is called the error or residual variance. From the perspective of a plant or animal breeder, specific environmental variance is basically random noise in the expression of the true phenotype, and it can interfere with artificial selection of traits of commercial interest by weakening the correlation between genotype and phenotype.
The second subcategory of environmental variance, general environmental variance, is attributed to the nongenetic sources of variation between individuals that are experienced by multiple individuals in a population. This is typically the largest component of variance in populations in natural conditions. While specific environmental variance is determined by the variation within replicated genetic lines, to obtain an estimate of general environmental variance, replicates of each of the genetic lines need to be assessed in each natural or experimental environment of interest (Fisher et al., 2004; Merilä et al., 2004). If just one environmental treatment is used, then all nongenetic sources of variation are due to the specific environment (i.e., the different microenvironments of individuals).
Estimates of general environmental variance can be determined by growing or raising the same set of genetic lines in all environments of interest. For example, water is required for all plants, but the amount that promotes maximum growth differs among both species and genetic lines of a species. Consider an experiment that grows 10 genetic lines of corn in each of three levels of soil moisture (low, medium, and high). On average, the genetic lines grow slower in the low and high moisture soils as compared to the medium moisture soil. Here, the changes in growth rate due to the different water treatments are equal to the general environmental variance. In Figure 1A (called a reaction norm), where the different lines represent different genotypes, the change in height (estimate of growth rate) associated with the different watering treatments illustrates the environmental variance.
Finally, the third subcategory of environmental variance—genotype by environment interaction—involves the unique or different responses of genetic lines to general environmental variation. To better understand this concept, once again consider the corn grown in different water levels. Among the 10 genetic lines under study, the response to water availability may not be parallel. For instance, some genetic lines may be less negatively affected by low or excessive moisture levels. Returning to Figure 1A, notice the similar response to greater water availability across all genotypes. In this case, there is no genotype by environment interaction, as all of the genotypes have a similar response to the same environmental change. However, in Figure 1B, the lines cross, which indicates that the different genotypes have different responses to an identical change in water availability. This differential response to environmental change (or genotype by environment interaction) can contribute to a species' ability to be successful in heterogeneous environments (Barrett et al., 2005).
Environmental Variance and Phenotypic Plasticity
Both general environmental variance and genotype by environment interaction are aspects of a characteristic known as phenotypic plasticity. Phenotypic plasticity is a change in the phenotype associated with a given genotype in response to different environmental conditions. Some researchers believe that genotype by environment interaction provides the best estimate of the potential for phenotypic plasticity to evolve. Other researchers argue that the correlation of genotype expression across two environments is a better indicator of this potential (Via & Hawthorne, 2005).
Phenotypic Variance: A Summary
In summary, the phenotypic differences among the individuals in a population can be attributed to both genetic and environmental sources. Determination of the underlying sources is essential for assessment of the potential of the population to evolve and adapt to heterogeneous or changing environments.
References and Recommended Reading
Barrett, R. D. H., et al. Experimental evolution of Pseudomonas fluorescens in simple and complex environments. American Naturalist 166, 470–480 (2005)
Etterson, J. R. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. I. Clinal patterns of selection along an environmental gradient in the Great Plains. Evolution 58, 1446–1458 (2004)
Falconer, D. S., & Mackay, T. C. F. Introduction to Quantitative Genetics (London, Longman, 1996)
Fisher, K., et al. Genetic and environmental sources of egg size variation in the butterfly Bicyclus anynana. Heredity 92, 163–169 (2004)
Gienapp, P., et al. Climate change and evolution: Disentangling environmental and genetic responses. Molecular Ecology 17, 167–178 (2008)
Lynch, M., & Walsh, B. Genetics and Analysis of Quantitative Traits (Sunderland, MA, Sinauer Associates, 1998)
Merilä, J., et al. Variation in the degree and costs of adaptive phenotypic plasticity among Rana temporaria populations. Journal of Evolutionary Biology 17, 1132–1140 (2004)
Mousseau, T. A., & Fox, C. W., eds. Maternal Effects as Adaptations (New York, Oxford University Press, 1998)
Saastamoinen, M. Heritability of dispersal rate and other life history traits in the Glanville fritillary butterfly. Heredity 100, 39–46 (2008)
Via, S., & Hawthorne, D. J. Back to the future: Genetic correlations, adaptation and speciation. Genetica 123, 147–156 (2005)
Waldmann, P. Additive and non-additive genetic architecture of two different-sized populations of Scabiosa canescens. Heredity 86, 648–657 (2001)



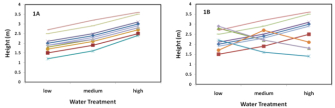
 Figure 1: The reaction norm.
Figure 1: The reaction norm.