« Prev Next »
The Beginnings of Molecular Ecology
As far back as the late 1800s, researchers realized that answers to some ecological questions could be obtained by examining the molecular composition of organisms. One of the earliest attempts at using molecules to address an ecological question was by Church in the late 1860s. Church studied relationships among birds and found that the pigment turacin was present only in birds of the Musophagidae family (Figure 1). He and others went on to determine that evolutionary relationships could be inferred according to whether species shared particular molecules. Early studies were limited to organic molecules obtained through diet and so may at times have confused relationships among organisms. However, the idea that the study of molecules could be a useful technique for understanding animals, their relationships, and their evolution had been firmly planted within the mind of the scientific community. And it is from this idea that the discipline of molecular ecology eventually emerged.

What exactly is molecular ecology? As we will see, it is an interdisciplinary approach to some of the most fundamental questions in organismal biology. Some scientists today do not consider it to be its own discipline but rather an "approach" taken in certain cases to answer certain questions. Most scientists, however, agree that it is distinct from other studies of organismal biology. Molecular ecology is defined as "the application of molecular techniques to answer ecological questions" (Beebee & Rowe 2004). In this article we explore the tools used in molecular ecology and how these tools enhance traditional ecological studies. We examine a number of seminal studies that have used molecular ecology tools. We also discuss the limitations of molecular ecology.
Tools of the Trade
One of the reasons why molecular ecology has advanced rapidly as a field of study is the advent of PCR. PCR makes it possible to amplify billions of copies of a specific piece of DNA from the genome with very few starting copies. In other words, it is possible to take a small sample of tissue to obtain enough DNA for study. This contrasts with earlier approaches that often required large amounts of DNA or protein, which often meant killing the organism of study. Obviously, killing one's study organism can be counter-productive, particularly if the intent of the study is to advance conservation efforts or protect endangered species. The fact that only a small starting amount of DNA is needed now for molecular ecology studies has opened the door for non-invasive sampling methods. It is now possible to isolate DNA from hair, urine, shed skin, and feces, thus preventing harm to endangered and non-endangered species. PCR also amplifies old and/or degraded DNA, such as that found in fossils (Figure 2).

Moving Beyond Traditional Ecological Approaches
Molecular techniques can be useful to, and sometimes necessary for, the field of ecology. Traditional approaches to ecology have limitations that can sometimes be addressed with molecular techniques. For one, traditional ecological approaches have relatively narrow timeframes of observation. Unless a long history of data has been directly collected on a particular organism through the years, traditional ecological approaches are limited to the period over which a study is conducted. Extrapolations can be made, but support for these extrapolations can be tenuous. On the other hand, historical events leave distinct signatures in the molecules of organisms that can be accurately interpreted.

A second important limitation of traditional ecological approaches is dependence on direct observation. One of the most commonly used methods of tracking animal movements and immigration into new populations is telemetry (Figure 5). Simply understanding an animal's physical movement from one place to another, however, often gives an incomplete picture of that animal's behavior and how it relates to its environment. One fundamental limitation of telemetry is that it cannot detect whether a dispersing animal successfully mates in its new territory or upon joining a new population. Reproductive success is an indication of fitness and, therefore, of the long-term survival of a population — a fundamental issue in ecology. But a DNA-based approach can provide much more insight into the mating behaviors of dispersing animals. By looking at molecular markers in a particular group of animals, researchers can establish the familial relationships among the members of that group; therefore, they can get an accurate picture of who is mating with whom. Recent immigrants will have slight differences in their molecular markers and they can be identified. If those immigrants are mating successfully in their new group, those differences will be passed onto their offspring and will appear more frequently. If immigrants do not mate, those differences will disappear.
Finally, since traditional ecological approaches are based on direct observations of organisms, they frequently do not detect underlying variation in organisms that does not influence physical appearances. It is possible for organisms to appear the same physically while exhibiting as much genetic divergence as found between distinct species. Cryptic variation can sometimes only be detected by comparing DNA.
These examples are intended to illustrate how molecular techniques can supplement and enhance information gained through traditional methodologies. We can get an even better idea of how molecular techniques have refined traditional ecology by looking at some of the fundamental questions answered by molecular ecology.
Answering Ecological Questions with Molecular Techniques
Some of the earliest molecular ecology studies involved examining the mating systems of birds. It was long thought that birds were monogamous. DNA samples of parents and their offspring challenged this idea. Indeed, it was found that extra-pair copulations were very frequent among bird species thought to be monogamous. As a result of the extensive use of DNA sampling of parents and their offspring, it is now understood that only a very small number of bird species adhere to a strictly monogamous mating system. Mating behaviors have also been described using molecular techniques in many other organisms including (among others) pipefish, frogs, beetles, and turtles.
Our understanding of habitat use has changed as the result of molecular techniques. Habitat use can be relatively simple to assess when animals can be tracked and directly observed using their habitat. But what happens when specific patterns of habitat use cannot be directly observed? There have been a number of studies on genetic structuring of populations that appeared to be uniformly distributed across a particular landscape. These studies found that populations were highly structured, indicating that organisms preferred to settle and mate in certain habitats over others. These findings contradicted observed distribution patterns at the landscape level because observational approaches were unable to track mating patterns.

Molecular approaches have also played a significant role in endangered species conservation besides addressing the question of cryptic variation. For example, molecular studies have have been used to identify migration corridors between populations that can prevent isolation of endangered populations. Molecular approaches have also been used to identify ideal populations for transfer to extinct or declining populations, methods for maximizing genetic variation in captive-bred animals, and species identification of contraband goods. Identifying species has been especially important for prosecuting poachers for harvesting endangered species when the only remains of the harvested animal is a piece of meat, bone, or fur. In fact, molecular techniques have been developed that can identify species based on the DNA from tissue even if the tissue has been cooked and mixed with other ingredients.
Limitations of Molecular Ecology
Molecular ecology has important limitations to consider. First, marker development can be time-consuming and expensive. Second, while it can be beneficial that molecular ecology is not dependent on direct observation of behaviors, this benefit can often be a limitation. Since the behavior is not directly observed, scientists must deduce the behavior that led to a specific molecular pattern, and there can often be multiple explanations for the same observed pattern. Third, it is not practical to look at the entire genome of all organisms, so one must look at a small subset of markers. Different markers may show discordant patterns or may not be representative of the entire genome. Finally, there are some questions that molecular ecology simply cannot answer and must be addressed with direct observation. For example, some behaviors important to the natural history of an organism, such as parental care and courtship behavior, can only be documented through direct observation.
References and Recommended Reading
Beebee T. & Rowe, G. An Introduction to Molecular Ecology, 2nd ed. New York, NY: Oxford University Press, 2008.
Beerli P., Hotz, H., et al. Geologically dated sea barriers calibrate an average protein clock in water frogs of the Aegean region. Evolution 50, 1676–1687 (1996).
Green D. M., Kaiser, H., et al. Cryptic species of spotted frogs, Rana pretiosa complex, in western North America. Copeia 1997, 1–8 (1997).
Highton R. Speciation in Eastern North American Salamanders of the Genus Plethodon. Annual Review of Ecology and Systematics 26, 579–600 (1995).
Highton R. Frequency of hybrids between introduced and native populations of the salamander Plethodon jordani in their first generation of sympatry. Herpetologica 54, 143–153 (1998).
Highton R. & Webster, T. P. Geographic protein divergence and variation in populations of the salamander Plethodon cinereus. Evolution 30, 33–45 (1976).
Moore M. K., Berniss, J. A., et al. Use of restriction fragment length polymorphisms to identify sea turtle eggs and cooked meats to species. Conservation Genetics 4, 95–103 (2003).
O'Brien, S. J., Wildt, D. E., et al. The cheetah is depauperate in genetic variation. Science 221, 459–462 (1983).
O'Brien, S.J., Roelke, M. E., et al. Genetic basis for species vulnerability in the cheetah. Science 227, 1428–1434 (1985).
Valenzuela N. Multiple paternity in side-neck turtles Podocnemis expansa: evidence from microsatellite DNA data. Molecular Ecology 9, 99–105 (2001).



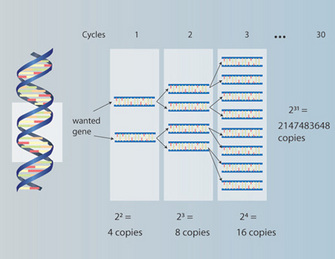
 Figure 2: Principle of polymerase chain reaction (PRC) to amplify target DNA sequence
Figure 2: Principle of polymerase chain reaction (PRC) to amplify target DNA sequence