Abstract
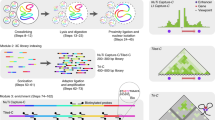
Genomic elements separated by large genomic distances can physically interact to mediate long-range gene regulation and other chromosomal processes. Interactions between genomic elements can be detected using the chromosome conformation capture (3C) technology. We recently developed a high-throughput adaptation of 3C, 3C-carbon copy (5C), that is used to measure networks of millions of chromatin interactions in parallel. As in 3C, cells are treated with formaldehyde to cross-link chromatin interactions. The chromatin is solubilized, digested with a restriction enzyme and ligated at low DNA concentration to promote intra-molecular ligation of cross-linked DNA fragments. Ligation products are subsequently purified to generate a 3C library. The 5C technology then employs highly multiplexed ligation-mediated amplification (LMA) to detect and amplify 3C ligation junctions. The resulting 5C library of ligated primers is analyzed using either microarray detection or ultra-high-throughput DNA sequencing. The 5C protocol described here can be completed in 13 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
References
ENCODE-consortium The ENCODE (ENCyclopedia Of DNA Elements) project. Science 306, 636–640 (2004).
Kleinjan, D.A. & van Heyningen, V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 (2005).
West, A.G. & Fraser, P. Remote control of gene transcription. Hum. Mol. Genet. 14, R101–R111 (2005).
Lomvardas, S. et al. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 (2006).
Dekker, J. A closer look at long-range chromosomal interactions. Trends Biochem. Sci. 28, 277–280 (2003).
de Laat, W. & Grosveld, F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11, 447–459 (2003).
Chambeyron, S. & Bickmore, W.A. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16, 256–262 (2004).
Dekker, J. The three C's of chromosome conformation capture: controls, controls, controls. Nat. Methods 3, 17–21 (2006).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Miele, A. et al. Mapping chromatin interactions by Chromosome Conformation Capture (3C). in Curr. Protoc. Mol. Biol. Vol. Suppl 74. (eds. Ausubel, F.M., Brent, R. Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. & Struhl, K.) 21.11.21–21.11-20 (John Wiley & Sons, Hoboken, New Jersey, 2006).
Splinter, E., Grosveld, F. & de Laat, W. 3C technology: analyzing the spatial organization of genomic loci in vivo . Methods Enzymol. 375, 493–507 (2004).
Tolhuis, B., Palstra, R.J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 (2002).
Palstra, R.J. et al. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 (2003).
Vakoc, C.R. et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 (2005).
Dostie, J. et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Spilianakis, C.G. & Flavell, R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5, 1017–1027 (2004).
Liu, Z. & Garrard, W.T. Long-range interactions between three transcriptional enhancers, active V kappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25, 3220–3231 (2005).
Murrell, A., Heeson, S. & Reik, W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 (2004).
Spilianakis, C.G., Lolioti, M.D., Town, T., Lee, G.R. & Flavell, R.A. Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645 (2005).
Kiermer, V. Story of C's. Nat. Methods 3, 872–873 (2006).
Hardenbol, P. et al. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 15, 269–275 (2005).
Wang, Y. et al. Allele quantification using molecular inversion probes (MIP). Nucleic Acids Res. 33, e183 (2005).
Yuen, T., Wurmbach, E., Pfeffer, R.L., Ebersole, R.J. & Sealfon, S.C. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30, e48 (2002).
Ling, J.Q. et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 (2006).
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 14, 477–495 (2006).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 (2006).
Gheldof, N., Tabuchi, T.M. & Dekker, J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc. Natl. Acad. Sci. USA 103, 12463–12468 (2006).
Rychlik, W., Spencer, W.J. & Rhoads, R.E. Optimization of the annealing temperature for DNA amplification in vitro . Nucleic Acids Res. 18, 6409–6412 (1990).
Breslauer, K.J., Frank, R., Blocker, H. & Marky, L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83, 3746–3750 (1986).
Ning, Z., Cox, A.J. & Mullikin, J.C. SSAHA: a fast search method for large DNA databases. Genome Res. 11, 1725–1729 (2001).
Singh-Gasson, S. et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17, 974–978 (1999).
Nuwaysir, E.F. et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12, 1749–1755 (2002).
Kim, T.H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005).
Selzer, R.R. et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 44, 305–319 (2005).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Osoegawa, K., de Jong, P.J., Frengen, E. & Ioannou, P.A. Construction of bacterial artificial chromosome (BAC/PAC) libraries. in Curr. Protoc. Mol. Biol. Vol. Suppl. 55. (eds. Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith & K. Struhl) 5.9.1–5.9.33 (John Wiley & Sons, Hoboken, New Jersey, 2001).
Nolan, T., Hands, R.E. & Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582 (2006).
Acknowledgements
This work was supported by a grant from the National Institutes of Health to J.D. (HG003143).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dostie, J., Dekker, J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc 2, 988–1002 (2007). https://doi.org/10.1038/nprot.2007.116
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.116
This article is cited by
-
Hi-Tag: a simple and efficient method for identifying protein-mediated long-range chromatin interactions with low cell numbers
Science China Life Sciences (2024)
-
Recent advances in chromosome capture techniques unraveling 3D genome architecture in germ cells, health, and disease
Functional & Integrative Genomics (2023)
-
Pattern recognition of topologically associating domains using deep learning
BMC Bioinformatics (2022)
-
Interplay of pericentromeric genome organization and chromatin landscape regulates the expression of Drosophila melanogaster heterochromatic genes
Epigenetics & Chromatin (2020)
-
Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection
Genome Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.