Abstract
Background:
Differentiation of systemic juvenile idiopathic arthritis (SJIA) fever from other childhood fevers is often delayed due to the lack of reliable, specific biomarkers. We hypothesized that PD-L1 expression is dysregulated in SJIA monocytes and compared it to other candidate SJIA biomarkers.
Methods:
This pilot study enrolled children with fever without source and compared PD-L1 expression on myeloid cells to C-reactive protein, erythrocyte sedimentation rate, leukocyte counts, S100A12, S100A8, S100A9, calprotectin, and procalcitonin. Logistic regression models were fit to test SJIA diagnosis with each marker used as an independent predictor. Receiver operating characteristic curves and area under curve were calculated. Gene expression profiling on a subset of samples was performed.
Results:
Twenty subjects (10 active SJIA, 10 febrile non-SJIA) were enrolled. S100 proteins were significantly elevated in SJIA with >80% sensitivity and >90% specificity. PD-L1 expression was significantly lower in SJIA. Other markers were not specific for SJIA. On exploratory gene analysis, 106 genes were significant for SJIA association, and several of these are associated with immune response pathways.
Conclusion:
In this small cohort, S100 proteins were specific diagnostic biomarkers for SJIA in children with fever. Decreased PD-L1 surface expression on circulating myeloid cells in SJIA suggests possible mechanism for loss of peripheral immune regulation.
Similar content being viewed by others
Main
Systemic juvenile idiopathic arthritis (SJIA) typically presents with prolonged fevers, arthritis, evanescent rash, lymphadenopathy, organomegaly, and/or serositis. SJIA diagnosis in clinical practice is fraught with difficulty due to the lack of a specific diagnostic test. Prior to initiating therapy, clinicians need to differentiate SJIA from other childhood causes of prolonged fever, such as infection, Kawasaki disease (KD), and malignancy. Identification of a biomarker to differentiate SJIA from other causes of childhood fever quickly and accurately would enable early targeted therapy (1), and shorter hospitalizations while decreasing patient and family suffering and morbidity. Distinguishing between SJIA and malignancy or infection is crucial in children, as corticosteroid therapy is often required to treat SJIA but is relatively contraindicated in children with underlying infections and can adversely affect the ultimate response to treatment and outcome in children with malignancy.
Attempts have been made to distinguish SJIA from its mimics, such as infection and malignancy (2,3). Usual markers of inflammation, such as C-reactive protein, erythrocyte sedimentation rate, and ferritin are not specific for SJIA. S100A12 (EN-RAGE), a calcium-binding protein secreted by activated phagocytes, is highly expressed in SJIA (specificity 94%, sensitivity 66%) (3) and KD (4), but not during infection or malignancy. Additionally, calprotectin (S100A8/9 heterodimer) is reported to be higher in patients with active SJIA compared with healthy controls, systemic infections, or malignancies (5,6). While elevated levels of IL-6 are associated with particular clinical features such as fever, thrombocytosis, C-reactive protein elevation, and osteoporosis (7), serum cytokine measurements are not helpful for SJIA diagnosis. Allantaz et al. (8) identified a leukocyte transcriptional signature that enables differentiation of SJIA from other febrile illnesses that awaits confirmation.
The pathogenesis of SJIA remains incompletely understood. The innate immune system is believed to play an important role. An expansion of cells of monocyte/macrophage lineage during SJIA flare (9,10,11), with increased expression of corresponding genes, has been reported (8,9,12,13,14). Crosstalk between the innate and adaptive immune system regulates a healthy inflammatory response. An important immunoregulator is programmed death ligand-1 (PD-L1), a glycoprotein expressed on antigen-presenting cells to limit T lymphocyte activation during an inflammatory response. Upon binding to its receptor, PD-L1 blocks T-cell proliferation and both IFN-γ and TNF-α production (15). PD-L1 expression is known to be upregulated during infection, malignancy, and most autoimmune diseases, including rheumatoid arthritis (16,17,18,19) and polyarticular JIA (unpublished data, Stevens Laboratory). In contrast, Mozaffarian et al. (20) have demonstrated decreased PD-L1 expression in systemic lupus erythematosus, suggesting a mechanism for loss of T lymphocyte regulation, and also a potential biomarker to distinguish SLE from infection. This pilot study tested the utility of PD-L1 expression as a biomarker to distinguish SJIA from other childhood febrile illnesses. We compared PD-L1 expression to other candidate markers for predictive value in SJIA diagnosis.
Results
Twenty-two subjects were enrolled in the study from 2010 to 2012. Two subjects, eventually diagnosed with mixed connective tissue disease and polyarticular JIA, were excluded. Eight subjects had newly diagnosed SJIA (treatment naive, four patients did not meet International League of Associations for Rheumatology criteria because they were treated with a presumptive diagnosis before 14 d of fever). Two subjects had known SJIA on immunosuppressive medications and presented with fever secondary to disease flare (subject 6 was on subcutaneous anakinra 2 mg/kg/d, oral prednisolone 0.03 mg/kg/d, and subcutaneous methotrexate 1 mg/kg/wk; subject 19 was on oral prednisolone 2 mg/kg/d, intravenous tocilizumab 12 mg/kg every 2 wk, and oral cyclosporine 4 mg/kg/d). The non-SJIA group comprised of six KD (two typical KD, of which one had coronary artery abnormalities; four with incomplete KD, of which two had coronary artery abnormalities), three infections, (viral lymphadenitis, pyelonephritis, and pneumonia), and one indeterminate. Subject 11 with incomplete KD and recent Epstein–Barr infection was classified as KD as this was believed to be the dominant cause of fever clinically (based on response to Intravenous immunoglobulin and initial echocardiogram demonstrating lack of distal tapering of coronary arteries).
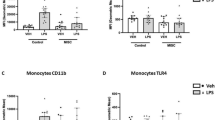
Table 1 depicts clinical and laboratory characteristics at enrollment. SJIA and non-SJIA groups differed only in incidence of arthralgia, arthritis, ALT, and neutrophil counts. We measured PD-L1 expression on myeloid cells by flow cytometry (Supplementary Figure S1 online) and found that PD-L1 expression on both monocytes (Mo) and activated monocytes (aMo) was significantly lower in SJIA ( Table 2 ). Subgroup analysis demonstrated that normalized PD-L1 mean fluorescence intensity expression was higher in both KD and infection compared to SJIA (median normalized mean fluorescence intensity PD-L1 Mo: SJIA 7, KD 50, infection 40, P = 0.009; aMo: SJIA 6, KD 56, infection 44, P = 0.07; myeloid dendritic cells (mDC): SJIA 5, KD 12, infection 11, P = 0.12).
The strongest association with SJIA was in S100 protein levels, all significantly higher in SJIA with area under curve (AUC) ≥ 0.95, and 80% sensitivity with 90% specificity for SJIA. Calprotectin was eightfold higher in SJIA with AUC 0.86 with 80% sensitivity and specificity (Supplementary Figures S2 and S3 online). Exploratory subgroups revealed significantly higher median S100 proteins in SJIA compared with KD or infection (S100A12 (ng/ml): SJIA 7,236, KD 316, infection 292, P = 0.003; S100A8 (ng/ml): SJIA 2,975, KD 214, infection 531, P = 0.006; S100A9 (ng/ml): SJIA 480, KD 44, infection 53, P = 0.005).
There were no differences in erythrocyte sedimentation rate or C-reactive protein between groups. Procalcitonin was elevated 10-fold in patients with infection but was not significantly different in SJIA (median procalcitonin (ng/ml): infection 2.5, SJIA 0.2, KD 0.7, P = 0.24). We were unable to determine the utility of ferritin as a biomarker as this was available for only two non-SJIA patients (median ferritin 482 ng/ml) and eight SJIA patients (median ferritin 971 ng/ml).
To identify novel biomarkers for SJIA, gene expression was explored by microarray. Principal component analysis, correlational heat map, and hierarchical clustering ( Figure 1 ) identified three subgroups with no clear association with disease status. All SJIA subjects except subject 21 and subject 19 clustered together. Subject 19 is a known SJIA patient on prednisone cyclosporine and tocilizumab. Expression data is publically available at NCBI GEO repository (21). Using a relaxed unadjusted P value of ≤0.001, 106 genes were significantly associated with SJIA. With one-way ANOVA false discovery rate at 0.05 (ref. 22), nine genes met criteria (BEX5, C11orf74, CTGF, LOC728368, PSPH, SNORD57, THUMPD3, ZC3H12B, and ZNF764). For functional analysis, we reduced significance criteria to unadjusted P value of ≤0.05 and applied an additional criterion of fold change >2 or < −2, compared to healthy controls. Finally, 1,161 probes met criteria and were uploaded to GeneGo Metacore (Thomson Reuters, New York, NY). The top 10 canonical pathways (Supplementary Tables S1–S3 online) were associated with cell cycle and immune responses. Interestingly, in contrast to surface expression detected by flow cytometry, PD-L1 mRNA expression was upregulated in SJIA.
Gene expression profiling in whole blood from patients with systemic juvenile idiopathic arthritis (SJIA), non-SJIA, and controls. (a) Principal component analysis result of 19,781 qualified probes. (b) Heatmap of hierarchical clustering results of correlation coefficients among sample pairs. (c) Principal component analysis result of 106 differentially expressed genes. (d) Heatmap of hierarchical clustering results of correlation coefficients among sample pairs. Figure key for panels a and c: A, infection; B, Kawasaki disease; C, control; D, SJIA. Figure key for panels b and d: KD, Kawasaki disease, iKd, incomplete Kawasaki disease.
Discussion
This exploratory study compares the utility of candidate biomarkers for their ability to distinguish SJIA from non-SJIA fever and provides the first data on PD-L1 and procalcitonin in SJIA. PD-L1, an immunoregulatory protein, is upregulated in rheumatoid arthritis (23), inflammatory bowel disease (24), and infection (16,17). Gene expression profiles may be affected by differing leukocyte composition in individual patients. Interestingly, although we noted decreased expression of PD-L1 surface protein in monocytic cells of SJIA, whole blood PD-L1 mRNA expression was upregulated, possibly due to the abundance of neutrophils in whole blood expressing PD-L1 (Stevens Laboratory, unpublished data and reported in Jarvis et al. (25)). SJIA subjects had significantly higher neutrophils compared to non-SJIA subjects. The impact of PD-L1 on neutrophils in controlling lymphocyte reactivity has not been explored.
In keeping with previous reports, our study confirms the utility of S100A12 as a diagnostic biomarker for SJIA (3,26). Our data additionally show that S100A12 may have potential to distinguish KD from SJIA, although replication of this finding in a larger cohort is essential. While the calprotectin complex has been shown to be more relevant in SJIA (5,6,27) with increased levels in synovium and serum, we show that S100A8 and S100A9 monomers are also increased in SJIA.
Procalcitonin was reported to have excellent sensitivity and specificity for bacterial fever in Still’s disease (28). Our data demonstrating elevated procalcitonin in infections is consistent with this, although the number of subjects with infection was low.
Expression array data overlapped with previous studies: 41 (including IL1B, NFATC3, STAT4 and other genes associated with the innate immune response), 2 (ATP2B1 and RNF14) and 64 genes were also identified by Ling et al., Allantaz et al., and the ANAJIS trial, respectively (8,29,30). Of the nine genes in our group that met the more stringent false discovery rate, interestingly ZC3H12B encodes a protein with a role in proinflammatory actions of macrophages (31). In addition, connective tissue growth factor (CTGF) is associated with several functions including fibrosis, angiogenesis, and tumorigenesis and is believed to play a role in inflammatory disorders including rheumatoid arthritis and CTGF polymorphisms are associated with systemic sclerosis (32,33,34).
Limitations of the study include small sample size, lack of patients with periodic fever or malignancy, sampling bias, and lack of a replication cohort. We were unable to assess the impact of immunosuppression on SJIA biomarkers, as only two patients were on such medications. Strengths of the study include the novel observation of decreased PD-L1 expression in SJIA that may point to an additional pathway important in pathogenesis and validation of S100A12 as a biomarker specific for SJIA.
To summarize, this pilot study determined that S100 proteins are indeed potential biomarkers to diagnose and distinguish SJIA fever from other causes of fever. We also determined that PD-L1 expression on SJIA myeloid cells was decreased unlike previously reported in patients with infectious diseases, a novel finding that requires replication in larger cohorts of SJIA subjects.
Methods
This study was approved by Seattle Children’s Hospital Institutional Review Board, and informed consent was obtained. All pediatric admissions, emergency department records, and outpatient rheumatology records were screened for eligible subjects. The study enrolled children 11 mo to 18 y with fever without source (fever without source was defined as: daily temperature ≥ 38 °C ≥ 5 d in a previously healthy child or daily fever ≥38 °C for ≥ 2 d in a child previously diagnosed with SJIA). Exclusion criteria were: (i) other rheumatic diseases; (ii) immunosuppressive therapy 2 wk prior to enrollment (including corticosteroids >0.5 mg/kg/d) except known SJIA patients were allowed to be on immunosuppressive therapy as long as there were no changes to their medications during the 4 wk prior to enrollment; (iii) pregnancy; (iv) hematocrit <20%; (v) stem cell or organ transplantation.
Blood was collected into Cell Preparation Tubes (BD Biosciences, San Jose, CA) and PAXgene RNA tubes (Qiagen, Valencia, CA). Peripheral blood mononuclear cells and plasma were isolated within 6 h and cryopreserved. PD-L1 expression assays were performed as previously reported (20,35). PD-L1+ cells segregated into monocyte (Mo: CD14highCD11c+), activated monocyte (aMo: CD14+CD11c+CD16+), and immature myeloid dendritic cell (mDC: CD14lowCD11c+) populations. To normalize for day-to-day variation in flow cytometer settings, mean fluorescence intensity for PD-L1 was normalized to background mean fluorescence intensity of the CD3–CD14–CD11c– subset (consistently negative for PD-L1 expression and similar to isotype controls) (Supplementary Figure S1 online).
Other biomarkers were analyzed in plasma by ELISA: procalcitonin (VIDAS B-R-A-H-M-S; BioMerieux, Durham, NC), S100 A12, A8, A9 (Circulex; MBL International, Woburn, MA), and Calprotectin (heterocomplex of S100 A8 and A9 proteins, Legend MAX Human MRP8/14; Biolegend, San Diego, CA).
Whole blood mRNA expression was analyzed by microarray on 14 samples (6 SJIA, 4 KD, 1 infection, and 3 healthy children). The Illumina Human HT-12 v4 Expression BeadChip (Illumina, San Diego, CA) was used with globin-reduced RNA (GlobinClear; Life Technologies, Grand Island, NY). RNA integrity was assessed using the Bioanalyzer 2100 (Agilent, Santa Clara, CA), and concentrations measured using NanoQuant (Tecan, San Jose, CA). cRNA was prepared by amplification with the Illumina TotalPrep RNA Amplification Kit (Life Technologies), hybridized, and scanned on HiScanSQ (Illumina).
Statistical Analysis
Tests were considered exploratory given small sample sizes. Final diagnosis was determined by chart review 12 wk after study enrollment. Subjects were categorized as: (i) SJIA by International League of Associations for Rheumatology criteria (36) or clinical diagnosis by attending pediatric rheumatologist (37); (ii) non-SJIA (infection, KD, indeterminate). The diagnosis of KD was made by attending pediatricians based on the 2004 American Heart Association algorithm for diagnosis and treatment of KD (38). Descriptive statistics were calculated including median, 25th, and 75th percentile for continuous variables and counts/proportions for categorical variables. With a relaxed α level = 0.1, Fisher’s exact test and exact Mann–Whitney rank-sum test were used to test differences in PD-L1 levels between groups. For adjusted analyses, values were log transformed. Multivariable logistic regression model was fit with SJIA indicator as response variable and each candidate marker as predictor, age, and gender as other predictors. Odds ratios estimated relative odds of SJIA diagnosis with unit increase in levels. Receiver operating characteristic curves to facilitate exploration of the sensitivity–specificity tradeoff at markers’ differing levels of predicted probability were generated. In addition, we report AUC adjusted for age and gender.
Microarray Statistics
GenomeStudio (Illumina) was utilized to generate background subtracted intensities and quality control metrics. 41.8% of 47,323 probes passed quality control and were processed by customized R/bioconductor pipeline, including quantile normalization (39), flooring, log2 transformation, and PALO filtering (present at least once; at least one sample must have had detection P value < 0.01). Analyses were performed using SAS 9.2 and JMP/Genomics 5.0 (SAS Institute, Cary, NC), functional analyses used GeneGo Metacore (Thompson Reuters, New York, NY).
Statement of Financial Support
S.S. was supported by the Early Career Investigator Award from the Thrasher Research Foundation, Salt Lake City, Utah, and National Institutes of Health (NIH; Bethesda, Maryland) T32 Training Award (AR7007108). REDCap was used for data management (UL1 RR025014 from National Center for Research Resources/NIH) through Seattle Children’s Research Institute and the Institute of Translational Health Science (ITHS), Seattle, Washington. E.D.M. was supported by NIH 1R21AR062765 and the University of California San Francisco-Stanford Arthritis Center of Excellence funded by the Great Western Region of the Arthritis Foundation.
Disclosure:
S.S., J.-N.O., V.H.G., C.N., C.M., E.D.M., and C.A.W. – none. A.M.S. – Patent for the use of PD-L1 as a diagnostic marker, licensed by Quest Diagnostics.
References
Vastert SJ, Kuis W, Grom AA. Systemic JIA: new developments in the understanding of the pathophysiology and therapy. Best Pract Res Clin Rheumatol 2009;23:655–64.
Jones OY, Spencer CH, Bowyer SL, Dent PB, Gottlieb BS, Rabinovich CE. A multicenter case-control study on predictive factors distinguishing childhood leukemia from juvenile rheumatoid arthritis. Pediatrics 2006;117:e840–4.
Wittkowski H, Frosch M, Wulffraat N, et al. S100A12 is a novel molecular marker differentiating systemic-onset juvenile idiopathic arthritis from other causes of fever of unknown origin. Arthritis Rheum 2008;58:3924–31.
Foell D, Ichida F, Vogl T, et al. S100A12 (EN-RAGE) in monitoring Kawasaki disease. Lancet 2003;361:1270–2.
Frosch M, Ahlmann M, Vogl T, et al. The myeloid-related proteins 8 and 14 complex, a novel ligand of toll-like receptor 4, and interleukin-1beta form a positive feedback mechanism in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum 2009;60:883–91.
Frosch M, Vogl T, Seeliger S, et al. Expression of myeloid-related proteins 8 and 14 in systemic-onset juvenile rheumatoid arthritis. Arthritis Rheum 2003;48:2622–6.
de Benedetti F, Massa M, Robbioni P, Ravelli A, Burgio GR, Martini A. Correlation of serum interleukin-6 levels with joint involvement and thrombocytosis in systemic juvenile rheumatoid arthritis. Arthritis Rheum 1991;34:1158–63.
Allantaz F, Chaussabel D, Stichweh D, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med 2007;204:2131–44.
Fall N, Barnes M, Thornton S, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum 2007;56:3793–804.
Hinze CH, Fall N, Thornton S, et al. Immature cell populations and an erythropoiesis gene-expression signature in systemic juvenile idiopathic arthritis: implications for pathogenesis. Arthritis Res Ther 2010;12:R123.
Macaubas C, Nguyen K, Deshpande C, et al. Distribution of circulating cells in systemic juvenile idiopathic arthritis across disease activity states. Clin Immunol 2010;134:206–16.
Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med 2005;201:1479–86.
Ogilvie EM, Khan A, Hubank M, Kellam P, Woo P. Specific gene expression profiles in systemic juvenile idiopathic arthritis. Arthritis Rheum 2007;56:1954–65.
Barnes MG, Grom AA, Thompson SD, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum 2009;60:2102–12.
Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD-1 pathway in the immune response. Am J Transplant 2012;12:2575–87.
Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol 2007;8:239–45.
Brown KE, Freeman GJ, Wherry EJ, Sharpe AH. Role of PD-1 in regulating acute infections. Curr Opin Immunol 2010;22:397–401.
Dong H, Strome SE, Matteson EL, et al. Costimulating aberrant T cell responses by B7-M1 autoantibodies in rheumatoid arthritis. J Clin Invest 2003;111:363–70.
Fife BT, Bluestone JA. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol Rev 2008;224:166–82.
Mozaffarian N, Wiedeman AE, Stevens AM. Active systemic lupus erythematosus is associated with failure of antigen-presenting cells to express programmed death ligand-1. Rheumatology (Oxford) 2008;47:1335–41.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;B57:289–300.
Wan B, Nie H, Liu A, et al. Aberrant regulation of synovial T cell activation by soluble costimulatory molecules in rheumatoid arthritis. J Immunol 2006;177:8844–50.
Nakazawa A, Dotan I, Brimnes J, et al. The expression and function of costimulatory molecules B7H and B7-H1 on colonic epithelial cells. Gastroenterology 2004;126:1347–57.
Jarvis JN, Petty HR, Tang Y, et al. Evidence for chronic, peripheral activation of neutrophils in polyarticular juvenile rheumatoid arthritis. Arthritis Res Ther 2006;8:R154.
Wittkowski H, Hirono K, Ichida F, et al. Acute Kawasaki disease is associated with reverse regulation of soluble receptor for advance glycation end products and its proinflammatory ligand S100A12. Arthritis Rheum 2007;56:4174–81.
Holzinger D, Frosch M, Kastrup A, et al. The Toll-like receptor 4 agonist MRP8/14 protein complex is a sensitive indicator for disease activity and predicts relapses in systemic-onset juvenile idiopathic arthritis. Ann Rheum Dis 2012;71:974–80.
Chen DY, Chen YM, Ho WL, Chen HH, Shen GH, Lan JL. Diagnostic value of procalcitonin for differentiation between bacterial infection and non-infectious inflammation in febrile patients with active adult-onset Still’s disease. Ann Rheum Dis 2009;68:1074–5.
Ling XB, Macaubas C, Alexander HC, et al. Correlation analyses of clinical and molecular findings identify candidate biological pathways in systemic juvenile idiopathic arthritis. BMC Med 2012;125:125.
Quartier P, Allantaz F, Cimaz R, et al. A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial). Ann Rheum Dis 2011;70:747–54.
Liang J, Wang J, Azfer A, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 2008;283:6337–46.
Liu SC, Hsu CJ, Chen HT, Tsou HK, Chuang SM, Tang CH. CTGF increases IL-6 expression in human synovial fibroblasts through integrin-dependent signaling pathway. PLoS One 2012;7:e51097.
Nozawa K, Fujishiro M, Kawasaki M, et al. Connective tissue growth factor promotes articular damage by increased osteoclastogenesis in patients with rheumatoid arthritis. Arthritis Res Ther 2009;11:R174.
Zhang X, Nie S, Si X, Luo Y, Tang W. Association between the CTGF -945C/G polymorphism and systemic sclerosis: a meta-analysis. Gene 2012;509:1–6.
Ou JN, Wiedeman AE, Stevens AM. TNF-α and TGF-β counter-regulate PD-L1 expression on monocytes in systemic lupus erythematosus. Sci Rep 2012;2:295.
Petty RE, Southwood TR, Manners P, et al.; International League of Associations for Rheumatology. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol 2004;31:390–2.
Behrens EM, Beukelman T, Gallo L, et al. Evaluation of the presentation of systemic onset juvenile rheumatoid arthritis: data from the Pennsylvania Systemic Onset Juvenile Arthritis Registry (PASOJAR). J Rheumatol 2008;35:343–8.
Newburger JW, Takahashi M, Gerber MA, et al.; Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease, Council on Cardiovascular Disease in the Young, American Heart Association. Pediatrics 2004;114:1708–33.
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–93.
Acknowledgements
We thank the patients and their families who participated in this study, Gretchen Henstorf for assisting with recruitment of patients, Matthew Crabtree and Dr. Rebecca Howsmon for technical assistance, Lael Agee for database management, Dr David Gilbert (Providence Portland Medical Center and Oregon Health Sciences University, Portland, OR) for performing procalcitonin assays, and Monica Chaudhari and Kathryn Whitlock for statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figure S1
(TIFF 24675 kb)
Supplementary Figure S2
(TIFF 8237 kb)
Supplementary Figure S3
(TIFF 33434 kb)
Supplementary Tables
(DOC 148 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Shenoi, S., Ou, JN., Ni, C. et al. Comparison of biomarkers for systemic juvenile idiopathic arthritis. Pediatr Res 78, 554–559 (2015). https://doi.org/10.1038/pr.2015.144
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pr.2015.144
This article is cited by
-
S100A8/A9, a potent serum and molecular imaging biomarker for synovial inflammation and joint destruction in seronegative experimental arthritis
Arthritis Research & Therapy (2016)