Abstract
Ultraconserved regions (UCRs) are >200 bp genomic segments with perfect human-to-rodent sequence identity. Transcribed UCRs constitute a new category of noncoding RNAs whose functions remain poorly understood. The human transformer 2β (TRA2B) gene contains a 419-bp UCR spanning the 276-bp exon 2 and its neighboring introns. TRA2B exon 2 has premature stop codons, whereas an exon 2-containing splice variant (TRA2β4) was expressed preferentially in the nuclei of human colon cancer cells. TRA2β4 knockdown p53-independently stimulated CDKN1A transcription and increased p21, resulting in the appearance of senescent cells. Biotin pull-down and RNA immunoprecipitation assays revealed that TRA2β4 interacted with Sp1 through a Sp1-binding sequence (485-GGGG-488) in a stem-loop structure of exon 2. Mutation of this sequence (485-AAGG-488) disrupted the stem-loop structure, blocked the interaction with Sp1 and increased CDKN1A transcription. Overexpression of TRA2β4 significantly decreased CDKN1A mRNA levels and accelerated cell growth, but the introduction of the mutation in the Sp1-binding sequence completely canceled these effects. Taken together, TRA2β4 may sequester Sp1 from occupying promoters of target genes including CDKN1A, promoting cell growth by interrupting the senescence-related gene expression program. This novel function of TRA2β4 may uncover an oncogenic function of transcribed UCRs.
Similar content being viewed by others
Introduction
Alternative splicing of pre-mRNAs generates protein diversity from a limited number of genes.1 The utilization of alternative splice sites is regulated by c is-acting elements and trans-acting factors through the interaction of a family of serine/arginine (SR)-rich splicing factors (SRSFs) and heterogeneous nuclear ribonucleoproteins. Alternative splicing occurs in a developmental stage-, sex- or tissue-specific manner and in response to the surrounding microenvironment.2, 3 High-throughput RNA sequencing of tissue-specific splicing events indicates that >90% of human genes produce multiple spliced isoforms.3 At the same time, aberrant alternative splicing participates in many genetic and acquired diseases including cancer.2, 4
An SR-like protein transformer 2β (Tra2β) regulates splice site selection of several genes encoding calcitonin/calcitonin gene-related peptide (C GRP), survival motor neuron 1 (SMN1) and microtubule-associated protein tau (TAU) in a concentration-dependent manner.5, 6, 7 Several lines of evidence suggest that overexpressed Tra2β may be involved in the pathogenesis of cancer.8, 9, 10 The human TRA2β (TRA2B) gene is composed of 10 exons and 9 introns. TRA2B contains a 419-bp genomic segment with perfect human-to-rodent sequence identity, termed the ultraconserved region (UCR; uc.138, see Supplementary Figure S1) spanning the 276-bp exon 2 and its neighboring introns.11, 12 TRA2B generates five mRNA isoforms (TRA2β1–5) through alternative splicing and usage of alternative promoters or polyadenylation sites (Figure 1a).12 Because of the existence of multiple premature termination codons (PTCs) in TRA2B exon 2, an exon 2-containing TRA2β4 mRNA isoform (hereafter referred as TRA2β4) is not translated and should be actively degraded through nonsense-mediated mRNA decay (NMD), a surveillance mechanism that degrades PTC-containing mRNAs. However, oxidative stress specifically facilitated TRA2β4 isoform production in rat gastric mucosa and a human gastric cancer cell line (AGS).13
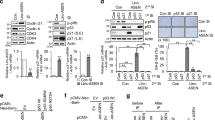
Expression of TRA2β1 and TRA2β4 in colon cancer cell lines. (a) Schematic diagram of the human TRA2B gene. Exons (ex) are indicated by open boxes and Arabic numbers. Filled boxes denote the ultraconserved exon 2. Five splice variants generated from TRA2B and the use of each exon are shown. (b) Amounts of TRA2β4 (upper panel) and TRA2β1 (lower panel) mRNAs in normal human colon and colon cancer cell lines (HCT116, RKO, HT29, Caco-2, T84 and SW480) were measured by qPCR using GAPDH mRNA as an endogenous quantity control. Values are means±s.d. from three independent experiments. *Significantly different by analysis of variance (ANOVA) and Bonferroni test (P<0.05). (c) HCT116 cells were treated with 10 μg/ml cycloheximide (CHX) for 4 h to inhibit NMD. Then, changes in mRNA levels of TRA2β1, TRA2β4, SRSF3 PTC and SRSF9 PTC were measured by qPCR using GAPDH mRNA as an endogenous quantity control. Values are means±s.d. from three independent experiments. (d) After treatment of HCT116 cells with 50 or 100 μg/ml CHX for 4 h, TRA2β4 levels were assayed by Northern hybridization with a locked nucleic acid (LNA) probe targeting for exon 2. 18S rRNA was used as a loading control. (e) After treatment of HCT116 cells with 10 nm of two different UPF1 siRNAs for 48 h, changes in TRA2β4 mRNA levels were measured by qPCR using GAPDH mRNA as an endogenous quantity control. Values are means±s.d. from three independent experiments.
Recently, it has become apparent that non-protein-coding RNAs (ncRNAs) are functionally important for normal development and physiology, as well as for pathologic processes.14 ncRNAs can be broadly classified into small (18–200 nt) and long ncRNAs (200 nt to >100 kb).15 Among them, epigenetic and genetic defects in a class of small ncRNAs called microRNAs are now recognized as a common hallmark of cancer. However, other ncRNAs, such as transcribed UCRs (T-UCRs), small nucleolar RNAs, PIWI-interacting RNAs or large intergenic noncoding RNAs, may also contribute to the development of many different diseases. There are 481 described UCRs, and more than half of them do not encode any protein.11 However, 68% of UCRs are transcribed, constituting a new category of ncRNAs, the T-UCRs.16 The wide distribution in the genome and lack of natural variation in the human population suggest an essential functional role in normal cells.11, 17 Recent genome-wide expression profiling studies have shown that certain T-UCRs are differentially expressed in human leukemias and carcinomas.16, 18, 19 Expression of those UCRs was suggested to be regulated by microRNAs, and the inhibition of an overexpressed T-UCR (uc. 73) induced apoptosis of colon cancer cells.16 T-UCRs are widely expressed in neuroblastomas and their expression correlates with important clinicogenetic parameters such as MYCN amplification status.20 However, the functional role of T-UCRs in cancer development is largely unknown.
Here, we introduce TRA2β4 as a functional T-UCR preferentially expressed in colon cancer cells. TRA2β4 may exert an oncogenic function by controlling senescence of colon cancer cells.
Results
Colon cancer cells upregulate TRA2β4 mRNA isoform expression
PTC-containing TRA2β4 mRNA isoform (TRA2β4) was considered to be degraded through NMD. However, when expression levels of TRA2β4 and TRA2β1 were examined in different colon cancer cell lines, all cell lines tested expressed significant amounts of TRA2β4 (Figure 1b, upper panel). TRA2β4 levels were estimated to be 10–20% of those of TRA2β1 mRNA that encodes a full-length Tra2β protein. On the other hand, TRA2β1 mRNA was not consistently overexpressed in the colon cancer cell lines (Figure 1b, lower panel). Human colon cancer (HCT116) cells expressed mainly TRA2β1 mRNA and increased TRA2β4 isoform production after exposure to sodium arsenite (Supplementary Figures S2b and c) as similarly observed in gastric cancer cells.13
Inactivation of NMD with cycloheximide increased the amounts of PTC-containing mRNA variants of SRSF3 and SRSF9 (Figure 1c). In contrast, cycloheximide treatment failed to increase TRA2β4 levels, which was confirmed by quantitative real-time reverse transcription– (qPCR) (Figure 1c) and Northern blot analysis (Figure 1d). Two different small interfering RNAs (siRNAs) targeting a key regulator of NMD (UPF1) effectively reduced UPF1 mRNA levels (Supplementary Figure S3a) and significantly increased the amounts of SRSF3 and SRSF9 PTC variants (Supplementary Figure S3b), but they did not increase TRA2β4 (Figure 1e).
To explain why NMD did not control TRA2β4 expression, we prepared nuclear and cytoplasmic fractions from HCT116 cells. The purity of each fraction was confirmed by western blotting using antibodies for cytosolic (α-tubulin) and nuclear (heterogeneous nuclear ribonucleoprotein C1/C2) marker proteins (Figure 2a), and by reverse transcription–PCR (RT–PCR) using primers targeting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) pre-mRNA (Figure 2b). RT–PCR amplified TRA2β4 in nuclear fractions containing GAPDH pre-mRNA, but not in cytosolic fractions, whereas TRA2β1 mRNA was distributed in both nuclear and cytoplasmic fractions (Figure 2b). qPCR measurement showed that TRA2β4 was significantly enriched in the nucleus, compared with TRA2β1 and GAPDH mRNAs (Figure 2c). RNA fluorescence in situ hybridization revealed that overexpressed TRA2β4 signals were present preferentially in the nuclei, whereas TRA2β1 mRNA signals were distributed in both the nucleus and the cytoplasm (Figure 2d). We also confirmed that HeLa cells constitutively expressed TRA2β4 in their nuclei similar to a nucleus-retained noncoding RNA (MEN ɛ/β) (Supplementary Figure S4a). Transiently overexpressed TRA2β4 was retained predominantly in the nuclei of HEK293T as well as of HeLa cells (Supplementary Figure S4b). These results suggest that significant amounts of TRA2β4 are retained in the nucleus and may escape from NMD-mediated degradation.
Subcellular distribution of TRA2β4 in HCT116 cells. (a) After nuclear and cytosolic fractions were prepared from HCT116 cells, the purity of each fraction was monitored by measuring a cytosolic protein (α-tubulin) and a nuclear protein (heterogeneous nuclear ribonucleoprotein (hnRNP) C1/C2) by western blotting. (b) Total RNA was extracted from each fraction and TRA2β1 mRNA or TRA2β4 was amplified by RT–PCR using specific primer sets. The purity of each faction was also assessed by RT–PCR measurement of GAPDH pre-mRNA. (c) TRA2β1, TRA2β4 and GAPDH mRNA levels in each fraction were measured by qPCR. Nuclear-to-cytosolic distribution ratio of each mRNA was calculated. Nuclear enrichment of TRA2β1 mRNA or TRA2β4 is indicated by comparing values to nuclear/cytosolic distribution ratio of GAPDH mRNA. Values are means±s.d. from three independent experiments. (d) Subcellular localization of TRA2β4 and TRA2β1 mRNA in HCT116 cells was examined by RNA fluorescence in situ hybridization (RNA-FISH) using locked nucleic acid (LNA) probes against exon 2 and exon 1–3 junction of TRA2β1 mRNA (green), respectively, as described in Materials and methods section. Cells were counterstained with TO-PRO-3 (blue). Scale bars, 5 μm.
TRA2β4 knockdown inhibits cell growth
To test the possibility that the TRA2β4 variant might be a functional T-UCR, we prepared two different siRNAs against TRA2β mRNAs (Tra2β siRNA targeting exon 6 and TRA2β4 siRNA targeting exon 2) and examined the effects of these siRNAs on cell growth. Although Tra2β siRNA was designed to degrade both TRA2β1 and TRA2β4 isoforms, it degraded TRA2β1 more effectively than TRA2β4 probably because of the predominant expression of the TRA2β1 isoform (Figure 3a). Consequently, Tra2β siRNA effectively reduced Tra2β protein (Figure 3b). In contrast, TRA2β4 siRNA selectively reduced TRA2β4 without changing TRA2β1 mRNA and Tra2β protein levels (Figures 3a and b). Treatment with Tra2β siRNA significantly inhibited cell growth (Figure 3c) in association with an increase in terminal deoxynucleotidyl transferase-mediated UTP end labeling (TUNEL)-positive cells (Figure 3d) and activation of poly(ADP-ribose) polymerase (PARP) and caspase-3 (Figure 3e). It was of interest that TRA2β4 siRNA did not change TRA2β1 mRNA or Tra2β protein levels, but it did inhibit cell growth (Figure 3c). Moreover, the selective TRA2β4 knockdown did not significantly increase TUNEL-positive cells (Figure 3d) and did not activate PARP and caspase-3 (Figure 3e).
Effect of TRA2β4 silencing on cell proliferation and apoptosis. (a) After HCT116 cells were treated with 10 nm of TRA2β4 or Tra2β siRNA for 48 h, TRA2β1 mRNA and TRA2β4 levels were measured by qPCR using GAPDH as an endogenous quantity control. Values are means±s.d. from six independent experiments. (b) HCT116 cells were treated with 10 nm TRA2β4, Tra2β or control siRNA for 48 h, and then the amounts of Tra2β were measured by western blotting using β-actin as a loading control. (c) HCT116 cells (1.5 × 104 cells) were seeded in 35-mm-diameter dishes and transfected with 10 nm TRA2β4, Tra2β or control siRNA. Subsequently, growing cells were harvested and counted at the indicated times. Values are means±s.d. from four independent experiments. *Significantly different by analysis of variance (ANOVA) and Bonferroni test (P<0.05). (d) After HCT116 cells were treated with 10 nm TRA2β4, Tra2β or control siRNA for 24 h, they were labeled using the DeadEnd Colorimetric TUNEL system (left panels), and the percentages of TUNEL-positive cells were determined (right panel). Values are means±s.d. from three independent experiments. *Significantly different by ANOVA and Bonferroni test (P<0.05). Scale bars, 50 μm. (e) After treatment of HCT116 cells with 10 nm TRA2β4, Tra2β or control siRNA for 48 h, whole-cell lysates were prepared from these cells. The levels of unprocessed or cleaved caspase-3 and PARP were measured by western blotting using β-actin as a loading control.
TRA2β4 knockdown induces cellular senescence
As shown in Figure 4a, TRA2β4 siRNA-treated HCT116 cells displayed unique morphological changes, that is, the cells spread out their cytoplasm and became thinner. These enlarged cells were positive for senescence-associated β-galactosidase (SA-β-gal) (Figure 4a). To confirm that TRA2β4 siRNA induced cellular senescence, we examined the expression of cyclin-dependent kinase inhibitors that have a crucial role in cell-cycle arrest and induction of senescence.21 Owing to a frameshift mutation in the INK4A gene and the hypermethylated promoter, p16 protein was undetectable in HCT116 cells.22 As shown in Figure 4b, treatment with TRA2β4 siRNA, but not with Tra2β1 siRNA, increased p21 levels. To avoid off-targeting effects, an additional siRNA targeting another sequence in TRA2β4 (TRA2β4 siRNA no. 2) were used. Transfection of TRA2β4 siRNA no. 2 also increased CDKN1A mRNA and p21 protein levels, and the percentage of SA-β-gal-positive cells (Supplementary Figure S5). It should be noted that TRA2β4 knockdown did not induce p53 (Figure 4b), and that TRA2β4 siRNA-mediated p21 induction occurred even in p53-null (p53−/−) HCT116 cells (Figure 4c). These results suggested that the reduction of TRA2β4 might increase CDKN1A mRNA and p21 protein levels in a p53-independent manner. We also confirmed that TRA2β4 knockdown similarly induced morphological changes and SA-β-gal in p53−/− HCT116 cells (Supplementary Figures S6a and b).
TRA2β4 knockdown induces cellular senescence. (a) HCT116 cells were transfected with 10 nm Tra2β, TRA2β4 or control siRNA for 72 h. They were then subjected to SA-β-gal staining. Scale bars, 10 μm. (b) After the transfection for 48 h, amounts of p21and p53 were measured by western blotting with respective antibodies (left panel). P21 levels were quantified using the Image J software (NIH, Bethesda, MD, USA) (right panel). β-Actin was used as a loading control. (c) After wild-type (p53+/+) or p53−/− HCT116 cells were treated with 10 nm TRA2β4 or control siRNAs for 48 h, CDKN1A mRNA levels were determined by qPCR using GAPDH mRNA as an endogenous quantity control. Data are expressed as fold changes compared with those in control siRNA-treated cells. Values are means±s.d. (n=5). (d) TRA2β4 levels in TIG cells at population doubling levels (PDL) 37, 40, 43, 53 and 62 were measured by qPCR using GAPDH mRNA as an endogenous quantity control. Data were expressed as fold changes relative to the levels in PDL 37 TIG-3 cells. Values are means±s.d. (n=3). (e and f) TRA2β4 and CDKN1A mRNA levels in young (PDL 37) and senescent (PDL 62) TIG-3 cells as well as young (PDL 29) and senescent (PDL 55) WI-38 cells were measured by qPCR using GAPDH mRNA as an endogenous quantity control. Values are means±s.d. from three independent experiments. *Significantly different by analysis of variance (ANOVA) and Bonferroni test (P<0.05) compared with those in control siRNA-treated cells.
A human fibroblast cell line (TIG-3) exhibited an age-associated reduction of TRA2β4 levels (Figure 4d). Reciprocal changes in TRA2β4 and CDKN1A mRNA expression were also observed along with senescence of TIG-3 and another human fibroblast cell line (WI-38); senescent cells decreased TRA2β4 expression in association with increased expression of CDKN1A mRNA (Figures 4e and f). These results suggest that TRA2β4 may regulate p21 expression during replicative senescence.
TRA2β4 interacts with Sp1 and regulates CDKN1A transcription
TRA2β4 knockdown did not affect the stability of CDKN1A mRNA (Supplementary Figure S7a) and p21 protein (Supplementary Figure S7b) or the translation rate of p21 (Supplementary Figure S7c), suggesting that the reduction of TRA2β4 may facilitate CDKN1A transcription. To confirm this, we cloned the 5′ flank of CDKN1A (from −2688 to +31 bp), and HCT116 cells were transfected with a luciferase reporter construct containing −2688/+31, −774/+31 or −163/+31 bp region of the human CDKN1A promoter (Figure 5a). The −2688/+31 bp region contained two p53-binding sites and exhibited basal and TRA2β4 siRNA-mediated upregulation of CDKN1A promoter activity (Figure 5a). The −163/+31 bp region as well as the −774/+31 bp region lost the p53-binding sites and markedly reduced the basal promoter activity. However, these two regions still responded to TRA2β4 knockdown and increased the luciferase activity. The −163/+31 bp region contained a cluster of Sp1-binding sites that likely mediated the response to TRA2β4 knockdown. To further investigate how reduction of TRA2β4 resulted in the stimulation of CDKN1A transcription, we performed chromatin immunoprecipitation assays with an anti-Sp1 antibody using control or TRA2β4 siRNA-treated HCT116. Sp1 did not bind to the -560/-226 region or the −325/+51 bp region in control siRNA-treated cells (Figure 5b), whereas in TRA2β4 siRNA-treated cells, Sp1 bound to the -325/+51 bp region containing the cluster of Sp1-binding sites, but not to the −560/−226 bp region (Figure 5b). These results suggest that Sp1 binding to the CDKN1A promoter may be blocked when significant amounts of TRA2β4 are present in the nucleus of HCT116 cells. The crucial role of Sp1 was also confirmed by two additional findings. That is, knockdown of Sp1 almost significantly blocked the TRA2β4 silencing-induced upregulation of CDKN1A promoter activity (Figure 5c), CDKNA1A mRNA expression (Figure 5d), and p21 protein expression (Figure 5e). In addition, co-transfection of Sp1 siRNA with TRA2β4 siRNA reduced TRA2β4-mediated increase in the percentage of SA-β-gal-positive cells (Figure 5d). Thus, the Sp1 elements seemed to play a pivotal role in the transcriptional activation of CDKN1A after silencing of TRA2β4.
TRA2β4 modifies promoter activity of the CDKN1A gene. (a) Twenty-four hours after transfection with 10 nm TRA2β4 or control siRNA, HCT116 cells were transiently transfected with luciferase reporter plasmids driven by −2688/+31, −774/+31 or −163/+31 bp promoter fragments of CDKN1A for 24 h. Luciferase activities in these cells were measured using the Dual-Luciferase Reporter Assay System. *Significantly decreased compared with control siRNA-treated cells (P<0.05 by analysis of variance (ANOVA) and Bonferroni test). (b) After treatment with TRA2β4 or control siRNA for 48 h, HCT116 cells were subjected to chromatin immunoprecipitation (ChIP) assays. Formaldehyde-crosslinked nuclear extracts were immunoprecipitated with an anti-Sp1 antibody or normal rabbit IgG (IgG). PCR was performed using an input nuclear chromatin fraction as a template (input). Specific PCR products corresponding to the region of the CDKN1A promoter containing the Sp1-binding sites were amplified and separated by agarose gel electrophoresis followed by ethidium bromide staining. (c) After treatment with 10 nm Sp1, TRA2β4 or control siRNA for 24 h, HCT116 cells were transiently transfected with the luciferase plasmid (pGL3-CDKN1A −163/+31) for 24 h. Luciferase activities in these cells were measured using the Dual-Luciferase Reporter Assay System. Values are means±s.d. (n=4). *Significantly different (P<0.05 by ANOVA and Bonferroni test). (d and e) After HCT116 cells were treated with TRA2β4 and/or Sp1 siRNA nos 1/2 as indicated for 24 h, expression levels of CDKN1A mRNA and p21 were analyzed by qPCR and western blotting. (f) After silencing of TRA2β4 and/or Sp1 nos 1/2, the cells were stained with SA-β-gal and then one hundred cells per individual sample in three independent fields were measured. *Significantly different (P<0.05 by ANOVA and Bonferroni test).
As shown in Figure 6a, RNA immunoprecipitation (RIP) assays indicated that an anti-Sp1 antibody retrieved endogenous TRA2β4 more effectively than TRA2β1 mRNA or other control RNAs, such as UBC, U6 and 18S, suggesting the association between Sp1 and TRA2β4. We prepared a plasmid encoding full-length TRA2β4 bearing bacteriophage MS2 hairpins (pMS-TRA2β4; Figure 6b). HCT116 cells were co-transfected with pMS-TRA2β4 and pMS2-yellow fluorescence protein (YFP) bearing an YFP-fused MS2-binding protein.23 Using this system, we again confirmed the association between MS2-tagged TRA2β4 and Sp1 in YFP-immunoprecipitated materials (Figure 6c). The presence of exon 2 in TRA2β4 is the only difference between TRA2β4 and TRA2β1, biotin-pull-down assays with HCT116 cells transcribing TRA2β exon 1, 2 or 3 RNA demonstrated the specific association of Sp1 with TRA2β exon 2 RNA (Figure 6d). As shown in Figure 6e, TRA2β4 exon 2 contains two Sp1 consensus sequences (485-GGGG-488 and 515-ACGG-518). CentroidFold (http://www.ncrna.org/centroidfold) and M-FOLD (http://mfold.rna.albany.edu/?q=mfold) programs indicate that TRA2β4 contains a stem-loop structure within exon 2 (449–488 nt, underlined in Figure 6e). Both programs also indicate that the introduction of two-point mutations at the Sp1-binding sequence (exon 2 mt-1, 485-GGGG-488 to 485-AAGG-488) completely disrupts the stem-loop structure (Figure 6e). The MS2 RNA hairpin-tagging system was again used to identify the interaction between exon 2 and Sp1. We constructed pMS2-LUC (Renilla luciferase gene (LUC))-exon 2 wild-type (exon 2 wt), pMS2-LUC-exon 2 mt-1 and pMS2-LUC-exon 2 mutated at another Sp1-binding sequence (exon 2 mt-2, 515-ACGG-518 to 515-ACAA-518) (Figure 6f) and transfected into HCT116 cells (Figure 6g). RIP between Sp1 and LUC-fused exon 2 constructs indicated that two-point mutations in the stem-loop motif (exon 2 mt-1), but not in another Sp1-binding site (exon 2 mt-2), significantly blocked the association between TRA2β4 exon 2 and Sp1 (Figure 6h), suggesting a crucial role of the consensus Sp1-binding site (485-GGGG-488) within the stem-loop motif.
TRA2β4 binds to Sp1 via exon 2. (a) Nuclear lysates prepared from UV-crosslinked HCT116 cells were subjected to an RIP assay using an anti-Sp1 antibody or normal mouse IgG. Immunoprecipitated RNAs were quantified by qPCR. Data are shown as enrichment relative to values obtained with normal mouse IgG. Values are means±s.d. (n=4). (b) Scheme for full-length TRA2β4 bearing bacteriophage MS2 hairpins (pMS-TRA2β4). YFP, yellow fluorescence protein. (c) After HCT116 cells were co-transfected with pMS2-YFP and pMS-TRA2β4 or control mock (pMS), association between Sp1 and TRA2β4 was analyzed using immunoprecipitaion with anti-YFP antibody and western blotting with anti-Sp1 antibody. (d) Nuclear fractions (40 μg) prepared from HCT116 cells were incubated with 1 μg of biotinylated transcripts designed as exon 1 (ex1), exon 2 (ex2) and exon 3 (ex3) of TRA2β4 in 10 mm Tris-HCl buffer, pH 8.0, containing 1 mm EDTA, 250 mm NaCl and 0.5% Triton X-100 for 1 h at room temperature. RNA–protein complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads, and bound Sp1 was detected by western blotting. (e) Nucleotide sequence of TRA2β4 exon 2. Two consensus Sp1-binding sites are boxed. Formation of stem-loop structure of TRA2β4 exon 2 RNA (449–488 nt) and its disruption by the mutation of 485-GGGG-488 to 485-AAGG-488 are shown below. These structures were predicted using CentroidFold and M-FOLD programs. (f) Scheme for pMS2-LUC (Renilla luciferase gene (LUC)), pMS2-LUC-exon 2 wild-type (pMS-ex2 wt), pMS2-LUC-exon 2 mt-1 (pMS-ex2mt-1), and pMS2-LUC-exon 2 mt-2 (pMS-ex2mt-2). Arrows indicate a primer set used. (g and h) Nuclear lysates prepared from UV-crosslinked HCT116 cells were subjected to an RIP assay using an anti-Sp1 antibody. Immunoprecipitated LUC-fused exon 2 RNAs were measured by qPCR. Data are shown as enrichment relative to values obtained from amount of each input. Values are means±s.d. (n=4).
We also examined whether the interaction between TRA2β4 and Sp1 was actually involved in regulating the expression of other Sp1-regulated genes, and found that TRA2β4 knockdown significantly increased the expression of KLF5, DRG2 and PRKRA mRNAs (Supplementary Figure S8). Sp1 is crucial for transcription of these genes.24, 25, 26
Expression of TRA2β4 in colon cancer cells promotes cell growth
To investigate the possible roles of TRA2β4 in human colon cancer, we generated a pCMV construct encoding TRA2β4 (pCMV-TRA2β4) or TRA2β4 containing the mutation (exon 2 mt-1, 485-GGGG-488 to 485-AAGG-488) in the stem-loop motif (pCMV-TRA2β4mt). Overexpression of TRA2β4 could significantly decease CDKN1A mRNA levels and accelerate cell growth, but the introduction of the mutation completely cancel these effects (Figures 7a and b). We also confirmed that the increased levels of TRA2β4 or TRA2β4mt did not affect TRA2β1 and p53 mRNA levels (Figure 7a).
Expression of TRA2β4 in colon cancers. (a) HCT116 cells were transfected with pcDNA3.1 (mock), pCMV-TRA2β4 or pCMV-TRA2β4 mutated in the stem-loop motif (pCMV-TRA2β4mt, 485-AAGG-488) for 48 h. mRNA levels were measured by qPCR using GAPDH as an endogenous quantity control. *Significantly decreased compared with mock-treated cells (P<0.05 by Student’s t-test). (b) After transfection of HCT116 cells (1 × 104 cells per 24-well dish) with pcDNA3.1 (mock), pCMV-TRA2β4 or pCMV-TRA2β4mt, the number of growing cells were measured using CellTiter96 AQueous Cell Proliferation Assay (MTS). Values expressed as means±s.d. (n=3). *Significantly increased compared with mock-transfected cells (P<0.05 by Student's t-test). (c) Using human colon cancer tissue qPCR arrays (TissueScan, HCRT103), TRA2β4 expressed in cDNAs from adenocarcinomas of the colon and surrounding normal colon tissues were measured by qPCR in 24 patients. Values were normalized to ACTB mRNA levels.
Finally, we measured the expression of TRA2β4 in 24 different cDNA libraries prepared from patients with colon cancer (Figure 7c). The relative expression of TRA2β4 varied depending on individual paired samples; however, colon cancer tissues expressed significantly higher levels of TRA2β4, compared with surrounding normal tissues.
Discussion
Among 481 UCRs discovered, 325 UCRs are transcribed and implicated to function as noncoding RNAs, whereas their specific functions are not fully investigated.16 We show here that human colon cancer cells express significant amounts of a T-UCR (TRA2β4), which is transcribed from uc 281. It has been shown that aberrant expression of specific T-UCRs is associated with chronic lymphocytic leukemia, colorectal cancer and hepatocellular carcinoma.16, 18, 19 We previously reported that Hu antigen R regulated alternative splicing of TRA2β to selectively produce TRA2β4 in human colon cancer cells under oxidative stress.27 Although TRA2β4 is a PTC variant, transcribed TRA2β4 was retained preferentially within the nucleus and resistant to the RNA surveillance NMD. Distinct ncRNAs retained within the mammalian cell nucleus are now referred to as nuclear-retained regulatory RNAs, and they are suggested to have structural roles or act as riboregulators.15, 28, 29 We suggest here that TRA2β4 may function as a novel regulator of senescence in colon cancer cells.
Tra2β is overexpressed in several types of cancers and has been suggested to participate in their abnormal growth.2, 8 In fact, our previous studies showed that Tra2β knockdown inhibited proliferation of colon cancer cells and induced their apoptotic cell death.9, 10 In the present study, we found that TRA2β4 (rather than Tra2β-encoding TRA2β1) was consistently overexpressed in all colon cancer cell lines tested. Although selective reduction of TRA2β4 inhibited cell growth, it did not stimulate apoptosis, but facilitated cellular senescence. These results suggest novel functions of TRA2β4 distinct from those of Tra2β protein. Cellular senescence is regulated by multiple factors, including p53, pRb and cyclin-dependent kinase inhibitors (p21, p16INK4a, p14ARF and p15INK4b). p16INK4a is one of the crucial factors for senescence in human tumors.30 However, like many other cancer cell lines, the p16INK4A gene is silenced in HCT116 cells owing to a frameshift mutation and the hypermethylated promoter.22 In contrast, TRA2β4 knockdown, but not Tra2β knockdown, induced p21 without changing p53 expression. The induction of p21 was observed even in p53−/− HCT116 cells. Upregulation of p21 induces senescence and the inactivation of this protein prevents senescence of colon cancer cells.31, 32 Thus, p21 was likely to be one of the key factors in the TRA2β4 knockdown-induced growth arrest and senescence of HCT116. Replicative senescence-dependent decline of TRA2β4 levels and a reciprocal induction of p21 observed in human fibroblast cell lines also support an important role of TRA2β4 in the regulation of cellular senescence. TRA2β4 knockdown could increase p21 levels in a p53-independent manner and it did not change either the stability or translation rate of CDKN1A mRNA. Based on the results, we speculated that TRA2β4 might directly regulate transcription of the CDKN1A gene, and found that TRA2β4 interacted with Sp1 via exon 2 and regulated CDKN1A mRNA expression. These results suggest that aberrantly expressed TRA2β4 may prevent Sp1 from occupying promoters of target genes, including CDKN1A, and thus promote cell survival by interrupting the senescence-related gene expression program. Sp1 is crucial for basal transcription of KLF5, DRG2 and PRKRA.24, 25, 26 We confirmed that TRA2β4 knockdown significantly increased the expression of these mRNAs in HCT116 cells, suggesting that interactions between TRA2β4 and Sp1 may be able to significantly alter the gene expression program.
Although the precise mechanism underlying the function of ncRNAs remains poorly understood, one emerging theme is the interaction between ncRNAs and protein complexes.33, 34 Several ncRNAs are required for the precise localization of chromatin proteins on genomic DNA targets.35 ncRNAs can also regulate the activity of protein complexes. An ncRNA upstream of CCND1 (ncRNACCND1) and the NRON ncRNA can bind to RNA-binding proteins or transcription factors and change their activities.36, 37 Distinct ncRNAs can work as molecular ‘decoys’. For example, the GAS5 ncRNA binds to the glucocorticoid receptor, blocking the correct binding to its regulatory elements.38 The long ncRNA PANDA interacts with the transcription factor NF-YA and limits the expression of proapoptotic genes, leading to apoptosis.39 At present, however, any T-UCR–protein interaction has not been documented.
TRA2β exon 2 has four purine-rich exonic splicing enhancers. Exonic splicing enhancer possesses an activity to retain RNAs in the nucleus through a saturable nuclear retention factor.40 Four purine-rich exonic splicing enhancers in TRA2β exon 2 may be important for nuclear retention of TRA2β4 to interact with Sp1 and other regulatory factors. In addition, RNA secondary structures, especially construction of stems, affected the RNA-protein-binding activity.41 Ultraconserved cDNA segments with AT-rich elements are resistant to secondary structure formation, keeping the segments open to allow regulatory factor binding.42 This would explain why Sp1 could bind preferentially to AT-rich exon 2 of TRA2β4. TRA2β4 exon 2 contains predictive Sp1-binding sites (485-GGGG-488 and 515-ACGG-518). CentroidFold and M-FOLD programs indicate the presence of a stem-loop structure (449–488 nt) within exon 2, which includes one Sp1-binding site (485-GGGG-488). Mutations of this site (485-AAGG-488) disrupted the stem-loop structure and eliminated the association between Sp1 and TRA2β4 exon 2. Thus, the consensus Sp1-binding site seems to be crucial for the interaction of TRA2β4 exon 2 with Sp1.
Overexpression of full-length TRA2β4 in HCT116 cells significantly reduced CDKN1A mRNA expression and accelerated their proliferation. The mutations (485-AAGG-488) completely eliminated these effects, again suggesting a crucial role of the consensus Sp1-binding site in the TRA2β4-mediated control of cell fate. Finally, we confirmed that colon cancer tissues expressed significantly higher amounts of TRA2β4.
The present study suggests that the TRA2B gene may control both apoptosis and senescence of colon cancer cells by generating different splice isoforms (TRA2β1 and TRA2β4). TRA2β4 RNA is likely to be a new anti-senescence factor working specifically in cancer cells. Cellular senescence is one of the important tumor-suppressive barriers.30, 31, 32, 43 It inhibits tumor cell proliferation and suppresses tumor cell motility, indicating that the induction of senescence results in the suppression of tumor cell growth, invasion and metastasis. Moreover, TRA2β4 knockdown could facilitate cellular senescence even in p53-deficient cells. Thus, TRA2β4 might be a novel tumorigenic ncRNA and a potential therapeutic target for colon cancer.
Materials and methods
Cell growth, apoptosis and senescence assays
Wild-type and p53−/− HCT 116 cells were cultured in McCoy’s 5A medium (Gibco, Grand Island, NY, USA) supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum and antibiotics at 37 °C in 5% CO2. T84 was maintained in Dulbecco's modified Eagle's medium/F-12 1:1 mixture (Gibco) supplemented with 10% fetal bovine serum. HEK293T, HT29, RKO and CaCo-2 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Human diploid fibroblasts (WI-38 and TIG-3) and SW480 cells were cultured in 10% fetal bovine serum-containing minimum essential medium. For the analysis of cell growth, appropriate numbers of HCT 116 or HEK293T cells were seeded in tissue culture plates, and the number of growing cells was counted using an automatic cell counter (Countess; Invitrogen, Carlsbad, CA, USA). Cell numbers on the plates were also assessed using the CellTiter96 AQueous Cell Proliferation Assay (MTS) (Promega, Madison, WI, USA). Apoptosis was evaluated by measuring cleaved caspase-9 and -3 levels by western blotting, and TUNEL analysis using the DeadEnd Colorimetric TUNEL system (Promega) according to the manufacturer’s protocol. Percentages of TUNEL-positive cells in five different fields (400 nm2 each) were calculated. The data were obtained in three independent experiments. Cellular senescence was assessed by the expression of SA-β-gal using an SA-β-gal Staining Kit (Cell Signaling Technology, Danvers, MA, USA).
Northern blot analysis
Locked nucleic acid-modified oligonucleotide probes targeting TRA2β transcripts (Gene Design, Osaka, Japan) were labeled with [32P]dCTP using recombinant terminal deoxynucleotidyl transferase (Life Technologies, Carlsbad, CA, USA). Oligonucleotide sequences using Northern blotting were listed in Supplementary Table S1. Samples of total RNA were separated in a 1% agarose gel containing 0.6 m formaldehyde and transferred to a nylon membrane filter. After prehybridization, the membrane was hybridized at 60 °C with the 32P-labeled probe overnight. After washing, bound probes were analyzed by BAS 1500 Image Analyzer (Fujifilm, Tokyo, Japan).
Preparation of nuclear and cytosolic fractions
After HCT116 cells were incubated in cytosolic lysis buffer (10 mm Tris-HCl, pH 7.4; 100 mm NaCl; 2.5 mm MgCl2; 40 μg/ml digitonin) for 10 min, lysates were centrifuged at 2060 g for 8 min at 4 °C, and supernatants were collected as cytosolic extracts. The remaining pellets were washed two times with the cytosolic lysis buffer and lysed with RIPA buffer (10 mm Tris-HCl, pH 7.4; 150 mm NaCl; 1 mm EDTA; 1 mm dithiothreitol; 0.1% sodium dodecyl sulfate; 1% Nonidet P-40). After centrifugation at 21 000 g for 10 min at 4 °C, supernatants were collected as nuclear extracts. Cells lysed with RIPA buffer were used as whole-cell extracts.
siRNAs
We used an siRNA (Hs_SFRS10_6 HP validated siRNA; Qiagen, Chatsworth, CA, USA) to knock down exon 3-containing TRA2β1 and TRA2β4 mRNAs. To silence selectively TRA2β4, exon 2-targeting siRNA was designed (Supplementary Table S1). A negative control siRNA (AllStars Negative Control siRNA) was obtained from Qiagen.
Quantitative real-time reverse transcription–PCR
Total RNAs were extracted from cells using TRIzol reagent (Life Technologies). One microgram of isolated RNA was reverse-transcribed using a PrimeScript RT Reagent Kit (Takara, Otsu, Japan). TRA2β1, TRA2β4 and CDKNA1 mRNA levels were measured using SYBR Green Master Mix and Applied Biosystems 7500 Real-time System (Applied Biosystems, Foster City, CA, USA). The sequences of primer sets are provided in Supplementary Table S1. TissueScan Tissue qPCR Arrays (HCRT103) including cDNAs from paired normal and tumor tissues in 24 patients with adenocarcinomas of the colon were obtained from OriGene Technologies (Rockville, MD, USA), and TRA2β4 levels in normal and tumor tissues were determined by qPCR. TRA2β4 levels were measured by the comparative ΔΔCt method using ACTB mRNA as a control and expressed as values relative to the normal samples.
Western blotting
Whole-cell lysates were prepared in a RIPA buffer (10 mm Tris-HCl, pH 7.4; 1% Nonidet P-40; 1 mm EDTA; 0.1% sodium dodecyl sulfate; 150 mm NaCl) containing a protease and phosphatase inhibitor cocktail (Roche Diagnostics Japan, Tokyo, Japan). Mouse monoclonal anti-α-tubulin (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Tra2β (1:1000; Abcam, Cambridge, UK), anti-caspase-3 (1:1000; Cell Signaling Technology, Danvers, MA, USA), anti-cleaved caspase-3 (1:1000; Cell Signaling Technology), anti-PARP (1:1000; Cell Signaling Technology), anti-cleaved PARP (1:1000; Cell Signaling Technology), anti-p21 (1:1000; Santa Cruz Biotechnology), anti-p53 (1:1000; Santa Cruz Biotechnology), anti-β-actin (1:1000; Abcam) or anti-Sp1 (1:1000, Santa Cruz Biotechnology) antibody was used.
Promoter activity assay
The 5′ flank of the human CDKNA1 gene was cloned into the pGL3-basic luciferase reporter vector (Promega). In brief, the first PCR was performed using human genomic DNA as a template. The CDKNA1 proximal promoter region was amplified using primer sets listed in Supplementary Table S1. The amplified products were subcloned into the pGL3-basic vector using HindIII and XhoI restriction sites. HCT116 cells (1.0 × 105) were cultured on 24-well plates, and then pGL3 luciferase constructs with various site-directed mutation or deletion (100 ng) were co-transfected with pRL-CMV vector (100 ng) using Jet-PEI (Polyplus Transfection, Illkirch, France). Twenty-four hours after the transfection, cells were harvested and the firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assays were performed using the Chromatin Immunoprecipitation Assay Kit (Millipore, Billerica, MA, USA). Briefly, HCT116 cells were fixed with 1% formaldehyde in phosphate-buffered saline for 10 min and then washed two times with ice-cold phosphate-buffered saline. These cells were resuspended in sodium dodecyl sulfate lysis buffer, incubated for 10 min on ice and then sonicated. Immunoprecipitation was carried out overnight at 4 °C using 3 μg antibody against Sp1. Normal rabbit IgG was used to assess nonspecific reactions. Immune complexes were collected with protein A agarose/salmon sperm DNA. Crosslinking between proteins and DNA was reversed according to the manufacturer’s protocol. Protein-bound DNA was extracted with phenol/chloroform/isoamyl alcohol. The extracted DNA was amplified by PCR (35 cycles; denaturing at 98 °C for 10 s, annealing at 55 °C for 30 s and extension at 72 °C for 1 min) using the following primers: for the CDKNA1A sequence between −560 and -226 bp, 5′-GGTGTCTAGGTGCTCCAGGT-3′ and 5′-GCACTCTCCAGGAGGACACA-3′; for the CDKNA1-B sequence between −325 and +51 bp, 5′-CAGCGCACCAACGCAGGCG-3′ and 5′-CAGCTCCGGCTCCACAAGGA-3′. The nuclear chromatin DNA from HCT116 cells (input) was used as a positive control for PCR.
Biotin pull-down assay
PCR fragments containing the T7 RNA polymerase promoter sequence were used as templates for in vitro transcription. Biotinylated transcripts were prepared by using a MEGAscript T7 Kit (Life Technologies) and biotin-CTP (Perkin-Elmer Japan, Yokohama, Japan), and purified with ssDNA/RNA Clean and ConcentratorTM (Zymo Research, Orange, CA, USA). Biotin pull-down assays were carried out by incubating 40 μg of nuclear fractions with 1 μg of biotinylated transcripts in TENT buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 250 mm NaCl and 0.5% Triton X-100) for 1 h at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Life Technologies), and bound proteins in the pull-down materials were analyzed by western blotting using an anti-Sp1 antibody.
RIP assay
HCT116 cells were lysed with 25 mm Tris-HCl buffer (pH 7.5) containing 150 mm NaCl, 1 mm EDTA, 1% (v/v) Nonidet P-40, 5% (v/v) glycerol and 100 U/ml RNase inhibitor (Promega). Whole-cell extracts (500 μg protein) were incubated with protein A Sepharose beads precoated with 3 μg anti-Sp1 antibody (Santa Cruz Biotechnology) or control rabbit IgG for 1 h at 4 °C. After washing with NT2 buffer (50 mm Tris-HCl buffer, pH 7.4, containing 150 mm NaCl2, 1 mm MgCl2 and 0.05% Nonidet P-40), beads were incubated with 20 U of RNase-free DNase I (Life Technologies) in NT2 buffer for 15 min at 37 °C and further incubated in NT2 buffer containing 0.1% sodium dodecyl sulfate and 0.5 mg/ml proteinase K for 20 min at 55 °C. RNA in the IP materials was measured by qPCR.2
Fluorescent in situ hybridization
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X for 5 min. RNA probes were prepared using Fluorescein Isothiocyanate RNA Labeling Kit (Roche Diagnostic, Mannheim, Germany) according to the manufacturer’s protocol. Then, cells were incubated for 16 h at 55 °C with 2 x SSC containing fluorescein isothiocyanate-labeled RNA probes. The cells were washed with 2 x SSC 55 °C for 30 min, the nuclei were stained using TO-PRO-3 (Life Technologies) and coverslipped with Vectashield (Life Technologies).
References
Modrek B, Lee C . A genomic view of alternative splicing. Nat Genet 2002; 30: 13–19.
Srebrow A, Kornblihtt AR . The connection between splicing and cancer. J Cell Sci 2006; 119: 2635–2641.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456: 470–476.
Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 2009; 16: 670–676.
Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K . Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells 2004; 9: 121–130.
Singh NN, Androphy EJ, Singh RN . In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 2004; 10: 1291–1305.
Tran Q, Coleman TP, Roesser JR . Human transformer 2beta and SRp55 interact with a calcitonin-specific splice enhancer. Biochim Biophys Acta 2003; 1625: 141–152.
Watermann DO, Tang Y, Zur Hausen A, Jäger M, Stamm S, Stickeler E . Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res 2006; 66: 4774–4780.
Kajita K, Kuwano Y, Kitamura N, Satake Y, Nishida K, Kurokawa K et al. Ets1 and heat shock factor 1 regulate transcription of the Transformer 2β gene in human colon cancer cells. J Gastroenterol 2013; 48: 1222–1233.
Kuwano Y, Nishida K, Kajita K, Satake Y, Akaike Y, Fujita K et al. Transformer 2β and miR-204 regulate apoptosis through competitive binding to 3′ UTR of BCL2 mRNA. Cell Death Differ 2015; 22: 815–825.
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS et al. Ultraconserved elements in the human genome. Science 2004; 304: 1321–1325.
Nayler O, Cap C, Stamm S . Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics 1998; 53: 191–202.
Takeo K, Kawai T, Nishida K, Masuda K, Teshima-Kondo S, Tanahashi T et al. Oxidative stress-induced alternative splicing of transformer 2beta (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am J Physiol Cell Physiol 2009; 297: C330–C338.
Cech TR, Steitz JA . The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157: 77–94.
Prasanth KV, Spector DL . Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev 2007; 21: 11–42.
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007; 12: 215–229.
Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK et al. Human genome ultraconserved elements are ultraselected. Science 2007; 317: 915.
Peng JC, Shen J, Ran ZH . Transcribed ultraconserved region in human cancers. RNA Biol 2013; 10: 1771–1777.
Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 2010; 29: 6390–6401.
Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010; 29: 3583–3592.
Lundberg AS, Hahn WC, Gupta P, Weinberg RA . Genes involved in senescence and immortalization. Curr Opin Cell Biol 2000; 12: 705–709.
Myöhänen SK, Baylin SB, Herman JG . Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res 1998; 58: 591–593.
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47: 648–655.
Chen C, Zhou Y, Zhou Z, Sun X, Otto KB, Uht RM et al. Regulation of KLF5 involves the Sp1 transcription factor in human epithelial cells. Gene 2004; 330: 133–142.
Fasciano S, Kaufman A, Patel RC . Expression of PACT is regulated by Sp1 transcription factor. Gene 2007; 388: 74–82.
Kim HJ, Ko MS, Kim HK, Cho WJ, Lee SH, Lee BJ et al. Transcription factor Sp1 regulates basal transcription of the human DRG2 gene. Biochim Biophys Acta 2011; 1809: 184–190.
Akaike Y, Masuda K, Kuwano Y, Nishida K, Kajita K, Kurokawa K et al. HuR regulates alternative splicing of the TRA2β gene in human colon cancer cells under oxidative stress. Mol Cell Biol 2014; 34: 2857–2873.
Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS . Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 2008; 105: 716–721.
Quinodoz S, Guttman M . Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol 2014; 24: 651–663.
te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP . DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res 2002; 62: 1876–1883.
Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV et al. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc Natl Acad Sci USA 2000; 97: 4291–4296.
de Carné Trécesson S, Guillemin Y, Bélanger A, Bernard AC, Preisser L, Ravon E et al. Escape from p21-mediated oncogene-induced senescence leads to cell dedifferentiation and dependence on anti-apoptotic Bcl-xL and MCL1 proteins. J Biol Chem 2011; 286: 12825–12838.
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477: 295–300.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–1323.
Guttman M, Rinn JL . Modular regulatory principles of large non-coding RNAs. Nature 2012; 482: 339–346.
Wang X, Arai S, Song X, Reichart D, Du K, Pascual G et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008; 454: 126–130.
Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005; 309: 1570–1573.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP . Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010; 3: ra8.
Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43: 621–629.
Taniguchi I, Masuyama K, Ohno M . Role of purine-rich exonic splicing enhancers in nuclear retention of pre-mRNAs. Proc Natl Acad Sci USA 2007; 104: 13684–13689.
Kwok CK, Tang Y, Assmann SM, Bevilacqua PC . The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci 2015; 40: 221–232.
Sathirapongsasuti JF, Sathira N, Suzuki Y, Huttenhower C, Sugano S . Ultraconserved cDNA segments in the human transcriptome exhibit resistance to folding and implicate function in translation and alternative splicing. Nucleic Acids Res 2011; 39: 1967–1979.
Badiola I, Santaolalla F, Garcia-Gallastegui P, Ana SR, Unda F, Ibarretxe G . Biomolecular bases of the senescence process and cancer. A new approach to oncological treatment linked to ageing. Ageing Res Rev 2015; 23 (Part B): 125–138.
Acknowledgements
This research was supported by a research grant (nos 26713027, 15K15294)) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to YK) and Takeda Science Foundation, Japan (to YK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Supplementary information
Rights and permissions
Oncogenesis is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kajita, K., Kuwano, Y., Satake, Y. et al. Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells. Oncogenesis 5, e213 (2016). https://doi.org/10.1038/oncsis.2016.18
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2016.18
This article is cited by
-
Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression
Scientific Reports (2021)
-
Accurate quantification of circular RNAs identifies extensive circular isoform switching events
Nature Communications (2020)
-
HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells
Scientific Reports (2019)