Abstract
Basal cell carcinoma (BCC) of the skin represents the most common malignancy in humans. MicroRNAs (miRNAs), small regulatory RNAs with pleiotropic function, are commonly misregulated in cancer. Here we identify miR-203, a miRNA abundantly and preferentially expressed in skin, to be downregulated in BCCs. We show that activation of the Hedgehog (HH) pathway, critically involved in the pathogenesis of BCCs, as well as the EGFR/MEK/ERK/c-JUN signaling pathway suppresses miR-203. We identify c-JUN, a key effector of the HH pathway, as a novel direct target for miR-203 in vivo. Further supporting the role of miR-203 as a tumor suppressor, in vivo delivery of miR-203 mimics in a BCC mouse model results in the reduction of tumor growth. Our results identify a regulatory circuit involving miR-203 and c-JUN, which provides functional control over basal cell proliferation and differentiation. We propose that miR-203 functions as a ‘bona fide’ tumor suppressor in BCC, whose suppressed expression contributes to oncogenic transformation via derepression of multiple stemness- and proliferation-related genes, and its overexpression could be of therapeutic value.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are ∼22 nt RNAs that regulate the expression of coding genes by binding imperfectly with their 3′-untranslated region (3′UTR), thereby causing target mRNA destabilization and degradation.1, 2 The ability of individual miRNAs to regulate hundreds of coding transcripts simultaneously enable these RNAs to control global gene expression programs and, ultimately, cellular physiology. Accordingly, biochemical and genetic studies have revealed that miRNAs function as key regulators in a wide variety of biological processes, including proliferation, differentiation, cell fate determination, apoptosis, signal transduction and organ development.3, 4 Moreover, the deregulation of miRNA expression contributes to human diseases, including developmental disorders, inflammation and cancer.5, 6, 7, 8
While tumors frequently overexpress specific ‘oncogenic’ miRNAs (oncomiRs, e.g., miR-155, miR-21 and miR-17∼92 miRs) that afford cancer cells with potent protumorigenic activity, tumor suppressor miRNAs are consistently downregulated in cancers (e.g., let-7, miR-16).7, 9 Deregulated expression of miRNAs has been associated with increased proliferation and migratory capacity, decreased apoptosis and a stem-cell-like phenotype, all prototypic features of cancer cells.10 Thus, modulation of miRNA activity represents an attractive therapeutic strategy that could be used in the treatment regimens of a majority of cancers.11, 12
Basal cell carcinoma (BCC) represents the most common malignancy in the Caucasian population, with a total of 1.3 million new cases in the year 2000 in the United States alone, posing a significant threat to public health.13, 14, 15 BCCs are keratinocyte tumors that histologically resemble proliferating epidermal progenitor cells derived from the basal cell layer of the interfollicular epidermis.13 Although the precursor cell from which BCC derives is thought to reside in the hair follicle, the precise cell-of-origin is still unclear.16, 17, 18, 19 The formation of BCCs is based on inherited factors combined with environmental factors, mainly, ultraviolet irradiation through long-term sun exposure. Family-based linkage studies of patients with basal cell nevus syndrome, a rare syndrome characterized by the susceptibility to develop multiple BCCs, identified mutations in the Patched 1 (PTCH1) gene, an inhibitor of the Hedgehog (HH) signaling pathway.13 The HH signaling pathway is currently thought to be of vital importance for the maintenance of cell growth in BCC.20, 21, 22, 23 Sporadic BCCs in human predominantly develop due to deregulation of HH pathway by inactivation of PTCH1 and subsequent activation of the GLI transcription factors.24 Despite recent advances in understanding the molecular alterations contributing to BCC, the pathogenesis is only partially understood. To date, all investigations on the onset and development of BCC have focused on mutations and/or expression of protein-coding genes, and a comprehensive molecular description detailing BCC pathogenesis is still lacking. At present, the role of miRNAs in the onset and progression of BCC is not known.
The work provided herein demonstrates that BCC tumors display a deregulated expression pattern of miRNAs compared with healthy human skin. We show that the ‘skin miRNA’ miR-203 is the most downregulated miRNA in BCCs and that overexpression of the c-JUN proto-oncogene, as well as activation of the HH and the epidermal growth factor receptor (EGFR) pathway may contribute to its reduced expression. We further demonstrate that miR-203 suppresses keratinocyte proliferation and directly targets c-JUN.
Results
MicroRNA expression profiling reveals major alterations in the BCC miRNAome
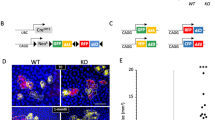
To explore the potential involvement of miRNAs in BCC, we compared the expression of 365 miRNAs in healthy skin and BCC. Using the Significance Analysis of Microarrays (SAM) algorithm, we identified 64 high-confidence, differentially expressed miRNAs in BCC relative to healthy skin that were significantly altered (false discovery rate: 2%; minimum fold-change: 2.0; Figure 1a and Supplementary Table 1). Unsupervised hierarchical clustering based on miRNA expression clearly separated BCC samples from healthy skin (Figure 1a). In accordance with previous reports relating to miRNA expression in solid tumors, the majority of differentially expressed miRNAs detected (62 out of 64) were suppressed in BCC (Figure 1a and Supplementary Table 1). These findings suggest that the altered expression of miRNAs may participate in the pathogenesis of BCC. The miRNA with the most significantly reduced expression in BCC was miR-203 (Supplementary Table 1), one of the most abundant miRNAs in skin (Supplementary Figure 1) that is preferentially expressed in keratinocytes and promotes epidermal differentiation by repressing stemness.25, 26
miR-203 is downregulated in BCC. (a) Unsupervised hierarchical clustering was performed on a subset of 64 genes that were differentially expressed between healthy skin (H) and basal cell carcinomas (BCC) as determined by significance analysis of microarrays. Heatmap colors represent relative miRNA expression. A median expression value equal to 1 was designated black; red, increased expression; and green, reduced expression. Note that the color scale is logarithmic (that is, 2 means fourfold change, 0 means no change). (b) Quantitative PCR analysis of the biologically active, mature form of miR-203 in healthy human skin (n=21) and BCC (n=22). The expression of miR-203 was normalized to U48 RNA. Data are expressed as arbitrary units. ***P<0.001, Mann–Whitney test. (c–d) In situ hybridization was performed on paraffin-embedded samples obtained from healthy skin and BCC using miR-203-specific locked nucleic acid (LNA) detection probes or scrambled LNA sequences. (c) Scoring was performed on a 0–6 scale, 0 indicating no discernible expression and 6 indicating strong expression in >80% of cells. ***P<0.001, Mann–Whitney test. (d) Representative image of miR-203 expression in healthy skin and BCC. Blue-purple color indicates miRNA expression.
Activation of the HH pathway results in suppressed mir-203 expression in BCC
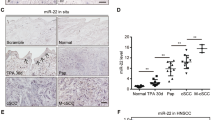
To validate our findings from the TaqMan MicroRNA Low Density Array (TLDA), we analyzed miR-203 levels in a larger number of samples obtained from healthy skin (n=21) and in BCCs (n=22) by quantitative real-time PCR (qPCR). In accordance with our previous data, miR-203 was present at high levels in healthy skin, while its expression was significantly reduced in BCCs (4.8-fold downregulation, P=8.5 × 10−9; Figure 1b).
Next, we performed in situ hybridization to visualize miR-203 expression in normal skin and BCC using specific locked nucleic acid probes designed to detect its abundance. In situ hybridization demonstrated that miR-203 was preferentially expressed in the suprabasal layers of healthy skin, while in BCCs, miR-203 expression was largely absent (Figure 1d), in line with our data obtained by qPCR. Moreover, scoring of in situ hybridization performed on a tissue microarray containing 8 healthy and 14 BCC samples showed a significant decrease of miR-203 expression in BCC (P<0.001, Figure 1c).
Upregulation of the HH pathway is currently assumed as the key molecular anomaly in all BCCs, and evidence suggests that aberrant activation of HH signaling is sufficient to initiate BCC carcinogenesis.27, 28 To explore the potential association between the downregulation of miR-203 and the activation of the HH pathway, we analyzed the expression of PTCH1 and the downstream HH effector GLI1 in BCC and healthy skin. Quantitative real-time PCR results showed that both PTCH1 and GLI1 were significantly overexpressed in BCC compared with healthy skin (P<0.001 for both, Figures 2a and c), in line with previously published data.13 A significant negative correlation was observed between miR-203 expression and GLI1 (P<0.0001, r=−0.81, Figure 2b), as well as between miR-203 and PTCH1 (P<0.0001, r=−0.74, Figure 2d), suggesting a causal relationship between HH activation and suppression of miR-203. To investigate whether activation of the HH pathway suppresses miR-203 in skin, we used a transgenic mouse model of BCC, in which human GLI1 is expressed in the skin under the control of the keratin 5 (K5) promoter (K5tTA/TREGLI1) in an inducible manner.29 These mice develop skin tumors closely resembling human BCCs and represent a highly relevant experimental model of spontaneous BCC having the same pathogenetic background as in humans.
Activation of the HH pathway suppresses miR-203 expression. Quantitative PCR analysis of Hedgehog genes GLI1 and PTCH1 in healthy human skin (n=18) and BCC (n=19). The expression of GLI1 (a, b) and PTCH1 (c, d) were normalized to 18S RNA. Data are expressed as arbitrary units. ***P<0.001, Mann–Whitney test. (b) and (d) Correlation of miR-203 expression with GLI1 and PTCH1 expression, respectively. MiR-203, GLI1 and PTCH1 expressions were measured by qPCR. Spearman correlation on log-transformed values. (e) Quantitative PCR analysis of miR-203 in wild-type (healthy) and K5tTA/TREGLI1 transgenic (BCC) mouse skin. (f) Detection of miR-203 in wild-type and K5tTA/TREGLI1 transgenic mouse skin by in situ hybridization.
To examine whether tumors in the K5tTA/TREGLI1 mice resemble human BCCs also in their miRNA expression pattern, we performed miRNA expression profiling in keratinocytes isolated from K5tTA/TREGLI1 and wild-type mice, using TLDA arrays. Overexpression of GLI1 resulted in the upregulation of 8 and the downregulation of 48 miRNAs in keratinocytes (Supplementary Table 2). Out of the 48 downregulated miRNAs, including miR-203, 15 have human orthologs that were also found to be significantly downregulated in human BCC (Supplementary Table 1). Thus, the miRNA profile of tumors developing in the K5tTA/TREGLI1 mouse model closely resembles that of human BCCs.
We further confirmed a marked decrease in the levels of miR-203 in keratinocytes isolated from the skin of K5tTA/TREGLI1 mice in comparison with wild-type animals by qPCR (Figure 2e). Moreover, in situ hybridization using specific locked nucleic acid probes for miR-203 demonstrated that miR-203 was expressed in the suprabasal epidermal layers both in wild-type and K5tTA/TREGLI1 mice, but lost in the basal layer and BCC-like lesions of the K5tTA/TREGLI1 animals (Figure 2f). These results indicate that activation of the HH pathway suppresses miR-203 expression in the skin in vivo. As the miR-203 promoter region does not contain any predicted GLI1 transcription factor binding sites, the suppression of miR-203 expression by the HH pathway is likely to be mediated by indirect mechanisms.
Activation of the EGFR pathway suppresses miR-203 expression
We have previously shown that miR-203 is regulated by the protein kinase C/activator protein 1 (AP-1) pathway and suppressed by growth factors such as keratinocyte growth factor and epidermal growth factor (EGF) in keratinocytes.25 Moreover, in silico transcription factor binding site analysis identified several AP-1 binding sites in the miR-203 promoter. Recently, it was shown that the EGFR signaling pathway synergizes with HH/GLI1 in oncogenic transformation via activation of the MEK/ERK/JUN pathway, indicating a possible mechanism for downregulation of miR-203 in BCCs.30, 31 To explore whether activation of the EGFR pathway is involved in human BCC, we analyzed the expression of EGF and EGFR in BCC and healthy skin. Immunohistochemical analyses confirmed that EGFR is expressed in BCC, as it is localized to the tumor cells (Figure 3a). Quantitative PCR results showed that both EGF and EGFR mRNA were upregulated in BCCs (n=9) as compared with healthy skin (n=10) (Figures 3b and c, P<0.01 for both). These results showed that activation of the EGFR signaling may have a role in a subset of BCCs.
Activation of the EGFR/MEK/ERK/JUN pathway suppresses miR-203 expression. (a) Immunohistochemical analysis of EGFR in healthy skin and human BCC. (b) Quantitative real-time PCR analysis of EGF in healthy skin (n=10) and human BCC (n=9); **P<0.01. (c) Quantitative real-time PCR analysis of EGFR in healthy skin (n=10) and human BCC (n=9); **P<0.01. (d) The expression of miR-203 in primary human keratinocytes cultured in the presence of dimethylsulfoxide vehicle control (CON), epidermal growth factor (EGF) alone, or in combination with inhibitors of MEK1/2 (UO126); JNK (SP600125); or of Akt (Wortmannin). The expression of the functionally active, mature form of miR-203 was analyzed using quantitative real-time PCR. Mean+s.d. of three independent experiments is shown. Expression data were normalized to U48 RNA; *P<0.05, **P<0.01. (e) Quantitative PCR analysis of miR-203 expression in c-JUN overexpressing keratinocytes; *P<0.05. (f) Schematic representation of the regulation of miR-203 by the EGFR/AP-1 and the Hedgehog pathway. n.s., nonsignificant.
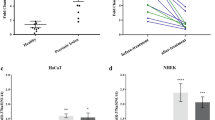
Therefore, we set out to further explore the role of EGFR signaling in the regulation of miR-203 by measuring its expression in primary human keratinocytes treated with inhibitors of MEK1/2 (UO126), JNK (SP600125) and Akt (Wortmannin), together with EGF or dimethylsulfoxide vehicle control (Figure 3d). Quantitative real-time PCR results showed a significant suppression of miR-203 in EGF-treated keratinocytes, which was reversed by SP600125 and UO126 (Figure 3d). By contrast, suppression of miR-203 by EGF was not reversed by the nonspecific Akt inhibitor Wortmannin, indicating that miR-203 is suppressed by the EGFR-mediated activation of the MEK/ERK but not by the PI3K/Akt pathway.
The oncoprotein c-JUN, part of the AP-1 transcription factor complex, has been shown to synergize with HH/GLI1 in oncogenic transformation via activation of the MEK/ERK/JUN pathway.30, 31 To investigate the involvement of c-JUN in the regulation of miR-203, we analyzed miR-203 expression in primary human keratinocytes transfected with a construct overexpressing c-JUN. Quantitative real-time PCR results showed that transient overexpression of c-JUN significantly suppressed miR-203 expression (Figure 3e, P<0.05), in accordance with our previous data in HaCaT cells.25
These results indicate that activation of the EGFR/MEK/ERK/JUN signaling pathway is a possible mechanism underlying reduced miR-203 expression in a subset of BCCs. A simplified scheme of the suggested mechanism of miR-203 regulation by the HH and EGFR signaling pathways is shown in Figure 3f.
c-JUN is a miR-203 target with deregulated expression in BCC
In order to gain further insight into the function of miR-203, we performed bioinformatic searches to identify putative miR-203-target interactions. Our previous analyses indicated that genes involved in the regulation of cell proliferation and cell cycle are particularly enriched among predicted miR-203 targets.32 Among several cell cycle–related targets, we identified c-JUN, a potent proto-oncogene commonly deregulated in a wide range of cancers,33 including skin tumors.34
To ascertain if miR-203 directly regulates c-JUN expression, we used luciferase reporter assays and demonstrated that c-JUN 3′UTR expression was significantly reduced (P<0.01) by miR-203 (pre-miR-203) co-transfection compared with scramble control (Figure 4a). Moreover, mutation in the predicted miR-203 binding site within the c-JUN 3′UTR relieved this reduction (Figure 4a), demonstrating a sequence-specific interaction between the miRNA and the c-JUN transcript. Furthermore, qPCR results showed that overexpression of miR-203 significantly suppressed c-JUN mRNA in primary keratinocytes (P<0.01, Figure 4b). In line with this, c-JUN protein levels diminished considerably in primary human keratinocytes 48 and 72 h after transfection with miR-203 precursors (Figure 4c).
miR-203 directly targets c-JUN. (a) Luciferase activity was measured 24 h after co-transfection of constructs containing the wild-type (WT) or mutant (MUT) 3′UTR of c-JUN gene together with transient overexpression of miR-203 or scramble miRNAs; **P<0.01. (b) qPCR analysis of c-JUN in primary human keratinocytes transfected with miR-203 precursor (pre-miR-203) or scramble control, or mock-transfected; **P<0.01. (c) Western blotting was used to analyze the expression of c-JUN protein in primary human keratinocytes 48 and 72 h after transfection with a pre-miR-203 or scrambled oligos. Tubulin served as a loading control. (d) Expression of miR-203, c-JUN and p63 in healthy human skin and in basal cell carcinoma were visualized by in situ hybridization and immunohistochemical staining, respectively.
Next, we performed immunohistochemical analysis of c-JUN protein in healthy human skin and in BCC. Similar to other miR-203 targets previously identified, such as p63,26 c-JUN was preferentially expressed in the basal, proliferative layer of healthy human epidermis. The suppression of miR-203 in BCC tumors was associated with a marked increase of c-JUN expression, evidenced by the intense and uniform distribution in BCC tumor nests (Figure 4d). Similar to c-JUN, p63, a key regulator of keratinocyte stemness, was also upregulated in BCCs (Figure 4d). Moreover, immunohistochemical analysis on the skin of K5tTA/TREGLI1 mice showed strong and uniform staining for both c-JUN and p63 in the BCC-like lesions of this model, similar to human BCC (Supplementary Figure 2). These results establish c-JUN as a novel target of miR-203 and show that loss of miR-203 may lead to derepression of its targets (c-JUN and p63) in BCC.
Overexpression of miR-203 reduces proliferation and causes a delay in G1- to S-phase transition
To further explore the functional role of miR-203 in BCC progression, we studied its effect on keratinocyte proliferation. To this end, we transiently overexpressed miR-203 by transfection of a synthetic precursor (pre-miR-203) into primary human keratinocytes and measured cell proliferation and cell cycle progression by EdU labeling and subsequent cell cycle analysis by flow cytometry (Figures 5a and b). Overexpression of miR-203 resulted in a fourfold reduction in the percentage of EdU-positive cells, in comparison with keratinocytes transfected with scrambled miRNAs (Figure 5b), indicating that miR-203 suppresses cell proliferation. More detailed analysis of the cell cycle distribution revealed that overexpression of miR-203 led to a significant increase in the percentage of cells in G1 phase and a decreased percentage of cells in S phase (Figure 5b). These results show that overexpression of miR-203 interferes with cell cycle progression from G1 to S phase. Hence, miR-203 acts to dampen mitogenic signals, presumably by targeting genes interconnected with keratinocyte-associated cell cycle progression and proliferation.35, 36
Overexpression of miR-203 suppresses keratinocyte proliferation by blocking G1- to S-phase cell cycle transition. (a) Primary human keratinocytes were transfected with a synthetic precursor molecule for miR-203 (pre-miR-203) or scrambled oligos as negative control (control, ctr). Cell proliferation and cell cycle progression was measured by EdU labeling and subsequent cell cycle analysis by fluorescence-activated cell sorting (FACS). Representative FACS plot from three independent experiments. (b) Bars depict mean±s.d. of the three independent experiments: percentage of cells in the G1, S and G2/M phase of the cell cycle and percentage of EdU+ cells are shown. ***P<0.001, Student's t-test.
Therapeutic delivery of miR-203 reduces the growth of BCC-like lesions in transgenic mice
We next investigated the effect of miR-203 overexpression on BCC growth in vivo. For this purpose, we used the K5tTA/TREGLI1 mouse model described above. To study the impact of miR-203 on the formation of BCC-like lesions, we administered miRIDIAN miR-203 mimics, or a negative non-targeting control, subcutaneously into mouse skin once every 48 h, starting 3 weeks after the induction of GLI1 (Figure 6a). Efficient delivery of miR-203 mimics was confirmed by qPCR (Supplementary Figure 3). Immunohistochemical analysis of SOX9, a marker for BCC and outer root sheath, which is not found in normal interfollicular epidermis, revealed robust morphological changes in mice treated with miR-203 mimics, with smaller number and decreased area of BCC-like lesions (Figure 6b). Moreover, overexpression of miR-203 led to decreased c-JUN and p63 expression (Figure 6c), indicating effective suppression of target genes in vivo by miR-203 mimics. These results show that therapeutic restoration of miR-203 interferes with BCC growth in vivo.
Subcutaneous administration of miR-203 mimics reduces BCC-like lesions in mice. (a) Experimental setup and timeline of the microRNA delivery experiments in the K5tTA/TREGLI1 mice. miRIDIAN miR-203 mimics (miR-203) or control nontargeting miRNA (Ctr) were delivered by subcutaneous injections. (b) Immunostaining of BCC-like lesions for SOX9, a general marker of BCC. Arrowheads indicate BCC-like lesions with SOX9-positive cell clusters. (c) Immunostaining of BCC-like lesions for the miR-203 targets c-JUN and p63 reveals decreased expression after delivery of miR-203 mimics.
Discussion
MiRNAs are important regulators of gene expression at the posttranscriptional level and are involved in basic biological processes, including cell proliferation, differentiation and apoptosis.3, 4, 37 Accordingly, deregulated miRNA expression has been implicated in diseases, such as cancer.3, 4, 8, 10 Here we show that miRNAs are deregulated in BCC and identify miR-203 as the most significantly and consistently downregulated miRNA. MiR-203 is an epidermal-specific miRNA, expressed preferentially in skin.26, 32 Moreover, of all the cellular constituents present in skin, miR-203 is predominantly expressed in keratinocytes, highlighting its importance in this cell type.32 Indeed, recent reports have shown that miR-203 is involved in skin morphogenesis, and promotes epidermal differentiation by repressing stemness of murine keratinocytes.38 We recently confirmed this finding in primary human keratinocytes, suggesting a conserved role for miR-203 function in the skin.25 Thus, low levels of miR-203 in BCC may be a prerequisite for tumor progression and the maintenance of an undifferentiated, stem-cell-like phenotype.
As miRNAs function as regulators of hundreds of distinct genes, the functional effects observed for miR-203 expression cannot be attributed to the regulation of a single target gene. SOCS-3 and p63 have previously been identified as miR-203 targets in keratinocytes.26, 32, 39 Our bioinformatic predictions suggested that genes involved in the regulation of cell proliferation are overrepresented among the targets of miR-203.32 Here we identify the c-JUN proto-oncogene as a novel target for miR-203, whose suppression may partially account for the observed reduction of cell proliferation by miR-203. c-JUN is a potent proto-oncogene commonly deregulated in a wide range of cancers,33 and has been implicated in the development of skin cancers.34 By virtue of its oncogenic origin, c-JUN has a crucial role in controlling homeostasis of cell growth. c-JUN belongs to the AP-1 family of transcription factors (which include protein members from the JUN, FOS, ATF and MAF families), which homo- or heterodimerize in order to exert a range of cellular and regulatory functions.33 By and large, the overall functional outcome of increased c-JUN activity is positive regulation of cell proliferation.40 Upon activation, c-JUN-containing AP-1 complexes induce transcription of positive regulators of cell cycle progression, such as cyclin D1,35, 41 while repressing negative regulators, such as p53 and INK4A. In accordance with the known regulatory role of c-JUN in cell transformation and cell cycle transition, we observed a miR-203-dependent block of the G1- to S-phase transition of the cell cycle, suggesting that suppression of c-JUN contributes to the observed antiproliferative effects of miR-203.
In agreement with our earlier results obtained with HaCaT cells,25 we demonstrate here that c-JUN suppresses miR-203 expression in primary keratinocytes. These data suggest the existence of a regulatory circuit, in which miR-203 and c-JUN mutually inhibit each other (Figure 7). This may represent a critical step in the regulation of basal cell fate commitment during differentiation. Keratinocyte differentiation is promoted upon the induction of miR-203 through the protein kinase C pathway,25 which in turn suppresses proliferative cues mediated by mitogenic proteins, such as c-JUN. As c-JUN is a negative regulator of miR-203, the circuit constitutes a feedback loop, whereby differentiating keratinocytes experience increasing levels of miR-203, resulting in a switch from proliferation to differentiation (Figure 7). Indeed, c-JUN mRNA levels have been shown to decrease significantly during human keratinocyte differentiation,42 while miR-203 is the most upregulated miRNA during this process.25 In support of this model, c-JUN in healthy skin is mainly expressed in the proliferative basal compartment of the interfollicular epidermis with little detectable immunoreactivity in differentiating suprabasal layers. In contrast, miR-203 expression is confined to suprabasal keratinocytes and is not expressed in the basal cell layer. Moreover, in BCC, suppressed miR-203 expression and function was accompanied by high c-JUN levels, with a level of expression similar to that of basal keratinocytes. This suggests that low levels of miR-203 contribute to the maintenance of a basal phenotype in BCC.
A simplified model of the interactions of miR-203, its regulators and targets in the context of BCC. The balance between miR-203 and its targets in undifferentiated cells can be shifted toward differentiation (under physiological conditions) and toward transformation (in BCC) by calcium (Ca2+)/protein kinase C (PKC) or Hedgehog (HH) signaling, respectively. The autoregulatory loop between c-JUN and miR-203 stabilizes the phenotype of cells once differentiation has been initiated, while lack of its upregulation allows sustaining an undifferentiated state with high proliferative capacity in BCC.
Indeed, c-JUN has recently been described as a major downstream effector of the HH pathway, whose direct activation by GLI in synergism with the EGFR/MEK/ERK/JUN pathway is essential for oncogenic activity of HH signaling in skin.30, 31 Our results showing that miR-203 is significantly suppressed by activation of the HH pathway as well as the EGFR pathway indicate an oncogenic mechanism by which suppression of miR-203 is sustained in transformed cells (Figure 7). Inappropriate activation of the HH and MAPK pathways in BCCs contributes to cancer progression via severely reduced expression of miR-203, which facilitates the misexpression of genes involved in the regulation of cell proliferation and cell cycle, including c-JUN, p63 and presumably other targets.
Taken together, our results indicate a complex molecular network, involving regulatory interactions between potent signaling pathways/oncoproteins and miR-203, which function in opposition to allow the precise spatial and temporal expression of genetic programs essential to basal and suprabasal proliferation and differentiation. The suppressed expression of miR-203 in BCC may thus cause the reiteration of molecular programs that normally govern skin development and homeostasis, ultimately leading to tumors that phenotypically resemble immature hair follicles.
In conclusion, we propose that miR-203 functions as a tumor suppressor miRNA in BCC. Similar to keratinocytes in the basal layer of the epidermis, where miR-203 expression is consistently low,32 severely reduced levels of miR-203 expression in BCCs is likely to sustain an undifferentiated phenotype with high proliferative capacity. Importantly, our results indicate that miR-203 along with its targets forms a regulatory circuit, which acts as a balance between proliferation and differentiation, under physiological conditions in the epidermis. In BCC, miR-203 functions as a downstream effector of the HH and the EGFR pathways, and its loss contributes to oncogenic transformation via derepression of multiple stemness- and proliferation-related genes. Finally, our results indicate that miRNA-203 may be a potential therapeutic target for the treatment of BCC.
Materials and methods
Clinical samples
Skin biopsies were taken, after obtaining informed consent, from healthy individuals and from patients with BCC, at the Department of Dermatology, Heinrich Heine University Dusseldorf, Germany, and at the Dermatology and Venerology Unit, Karolinska University Hospital, Stockholm, Sweden. The clinical diagnosis was made by a dermatologist and was confirmed by histopathological evaluation. All studies were approved by the Regional Committees of Ethics. Isolation of total RNA was performed as described.43 Skin cancer tissue array SK801 was purchased from US Biomax (Rockville, MD, USA).
MicroRNA expression profiling
MiRNAs were reverse transcribed and amplified (PCR) using the multiplex RT TaqMan MicroRNA Low Density Array (Applied Biosystems, Foster City, CA, USA). TLDAs were run on the ABI7900 HT analyzer with TLDA upgrade and analyzed with RQ Manager software provided by Applied Biosystems. All the quality control tests were validated: blanks and reproducibility (standard deviation of cycle threshold (Ct) <1) of the two small nucleolar housekeeping RNAs RNU48 (SNORD48) and RNU44 (SNORD44). The amount of RNA from each sample was calibrated to the more stable (between the different arrays) housekeeping RNA, RNU48. To find consistently differentially expressed genes, the data were subjected to significance analysis of microarrays as described.42 Genes showing at least twofold regulation and a q-value <2% were considered to be differentially expressed.
Quantitative real-time PCR
Quantification of miRNAs by TaqMan Real-Time PCR was carried out as described previously.42 Target gene expression was normalized between different samples based on the values of U48 small nucleolar RNA. Quantification of mRNAs was carried out according to standard procedures.19 Specific primers and probes were obtained from Applied Biosystems, except for 18S (18S-F: 5′-CGGCTACCACATCCAAGGAA-3′, 18S-R: 5′-GCTGGAATTACCGCGGCT-3′, 18S TaqMan probe: 5′-FAM/TGCTGGCACCAGACTTGCCCTC-3′).
In situ hybridization
In situ hybridization analyses for miR-203 were performed as described.42 Briefly, sections were fixed in 4% paraformaldehyde for 10 min, acetylated for 10 min, washed, and prehybridized for 1 h at 46 °C. Hybridization with digoxygenin-labeled miRCURY locked nucleic acid probes (Exiqon, Vedbaek, Denmark) was performed for 1 h at 45 °C. Slides were then washed at 45 °C and incubated with horseradish peroxidase–conjugated sheep anti-DIG Fab fragments (1:1000; Roche, Mannheim, Germany) for 30 min at room temperature. Sections were visualized by using BM purple substrate together with 2 mM levamisole (Vector Laboratories, Burlingame, CA, USA). The color reaction was performed over night at room temperature. Scoring of the in situ hybridization on the tissue array was performed on a scale 0–6 (intensity: negative: 0, weak: 1, moderate: 2, strong: 3, percentage of positive cells: 0%, 0, 1–25%, 1; 26–50%, 2; 51–100%, 3).
Transfections
Human adult skin epidermal keratinocytes (obtained from Cascade Biologics, Portland, OR, USA) were cultured in EpiLife serum-free keratinocyte growth medium, including Human Keratinocyte Growth Supplement at a final Ca2+ concentration of 0.06 mM (Cascade Biologics). Third passage keratinocytes at 70% confluence were transfected with 10 nM Pre-miR 203 miRNA Precursor (Ambion Applied Biosystems, Austin, TX, USA) or 10 nM Pre-miR miRNA Precursor Negative Control #1 (Ambion) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), following the manufacturer's instruction. The c-JUN cDNA44 was cloned into the CSII-CMV-MCS-IRES2-Bsd vector and transiently transfected into human keratinocytes using Fugene6 (Roche), according to the manufacturer's instructions.
Immunoblots
Keratinocyte lysates were analyzed for protein expression by western blotting with anti-human c-JUN antibody at a concentration of 1:500 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The protein levels were visualized by enhanced chemiluminescence (GE Healthcare, Niskayuna, NY, USA) using horseradish peroxidase–conjugated anti-rabbit antibody at a concentration of 1:2000 (Cell Signaling Technology, Beverly, MA, USA).
Growth factors and inhibitors
Primary human keratinocytes were treated with 10 ng/ml EGF (Sigma, St Louis, MO, USA) in the presence of various inhibitors or dimethylsulfoxide. c-JUN NH2-terminal kinase-inhibitor SP600125 (Santa Cruz Biotechnology), mitogen-activated protein kinase kinase 1 and 2 (MEK1/2) inhibitor, UO126 (Calbiochem, Darmstadt, Germany), and Akt inhibitor, Wortmannin (Calbiochem), were applied to primary human keratinocytes at the following concentrations: SP600125 (10 μM), UO126 (10 μM) and Wortmannin (1 μM).
3′UTR luciferase assays
Luciferase reporter plasmids containing the wild-type 3′UTR and the 3′UTR with mutated miR-203 binding site of c-JUN were obtained from Genecopoeia (Rockville, MD, USA) and Genscript (Piscataway, NJ, USA), respectively. HEK293 cells were transfected with the luciferase constructs (100 ng per 24-well) and pre-miR-203 or pre-miR-Control (10 nM, Applied Biosystems) using Lipofectamin 2000 (Invitrogen). Luciferase activity was measured after 24 h using Dual Luciferase Reporter Assay according to the manufacturer's instructions (Promega, Madison, WI, USA).
Immunohistochemistry
Paraffin-embedded normal skin and BCC sections (5 μm thickness) were baked at 60 °C for 1 h. Sections were deparaffinized in 1 × Aqua de Par (Biocare Medical, Walnut Creek, CA, USA) at 80 °C for 10 min. Slides were then transferred to 1 × DIVA decloaker reagent or 1 × EDTA decloaker reagent (Biocare Medical) and antigen retrieval was performed using a pressure boiler (Biocare Medical) as heat source. Sections were washed in Tris-buffered saline, blocked in peroxidaze 1 for 10 min, washed again, pretreated for 10 min in Background Sniper (Biocare Medical) and incubated with the following primary antibodies at 4 °C overnight: c-JUN (1:100, clone E254, Epitomics Burlingame, CA, USA); and p63 (1:500, clone BC4A4, Santa Cruz Biotechnology), EGFR (ready to use, clone 31G7; Invitrogen, Camarillo, CA, USA). After washing in TBS, slides were incubated with labeled polymer (mouse or rabbit) for 10 min using the MACH 3 AP-Polymer Detection Kit (Biocare Medical). Antibody staining was visualized using the Vulcan Fast Red Chromogen Kit (Biocare Medical) and the slides were counterstained with Tacha's Automated Hematoxylin (Biocare Medical).
Cell cycle analysis
Click-iT EdU Flow Cytometry Assay (Invitrogen) was carried out according to the manufacturer's instructions, and analyzed by flow cytometry on a FACScan (Becton Dickinson, Franklin Lakes, NJ, USA).
Mouse model of BCC
The mouse model of human BCC was generated by crossing mice expressing tetracycline-dependent transactivator under control of bovine cytokeratine 5 promoter (K5tTA) and mice harboring human GLI1 transgene under control of tetracycline response element (TREGLI1).29 GLI1 expression was induced by withdrawal of doxycycline (2 mg/ml) from the drinking water at postnatal day 16. Mouse keratinocytes were purified as described.44 Total RNA was extracted by RNA-Bee (TEL-TEST, Friendswood, TX, USA). After DNase treatment (Qiagen, Valencia, CA, USA), RNAs including short RNAs were isolated by RNeasy kit (Qiagen) according to the manufacturer's instructions.
In vivo delivery of miRNA mimics
MiRIDIAN miR-203 mimics and MiRIDIAN negative control sequence based on cel-miR-67 (Dharmacon, Lafayette, CO, USA) was complexed with in vivo-jetPEI (Polyplus Transfection, Strasbourg, France) in 400 μl of 5% glucose solution and injected subcutaneously into dorsal skin of K5tTA/TREGLI1 mice (n=4 each), once in every 48 h, starting at P35, 3 weeks after induction of GLI1 expression. The protocol was approved by the local ethics committee. At p58, 48 h after the last injection, mice were killed, skin samples were collected and snap-frozen for RNA isolation or paraffin-embedded for immunohistochemical staining. Successful delivery of the miRNA mimics was confirmed by qPCR (Supplementary Figure 3). SOX9 staining was performed, as described,29 on sections from skin stripes including the site of injection and adjacent skin.
Statistics
Statistical significance for experiments was determined by Mann–Whitney U-test or Student's t-test. Correlation between the expressions of different genes in the same samples was analyzed using Spearman correlation test on log-transformed data.
References
Filipowicz W, Bhattacharyya SN, Sonenberg N . Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114.
Guo H, Ingolia NT, Weissman JS, Bartel DP . Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466: 835–840.
Alvarez-Garcia I, Miska EA . MicroRNA functions in animal development and human disease. Development 2005; 132: 4653–4662.
Miska EA . How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 2005; 15: 563–568.
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635–647.
Makunin IV, Pheasant M, Simons C, Mattick JS . Orthologous microRNA genes are located in cancer-associated genomic regions in human and mouse. PLoS ONE 2007; 2: e1133.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Zhang B, Pan X, Cobb GP, Anderson TA . microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302: 1–12.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Croce CM, Calin GA . miRNAs, cancer, and stem cell division. Cell 2005; 122: 6–7.
Czech MP . MicroRNAs as therapeutic targets. N Engl J Med 2006; 354: 1194–1195.
Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S . The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol 2008; 18: 89–102.
Epstein EH . Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008; 8: 743–754.
Housman TS, Feldman SR, Williford PM, Fleischer Jr AB, Goldman ND, Acostamadiedo JM et al. Skin cancer is among the most costly of all cancers to treat for the Medicare population. J Am Acad Dermatol 2003; 48: 425–429.
John Chen G, Yelverton CB, Polisetty SS, Housman TS, Williford PM, Teuschler HV et al. Treatment patterns and cost of nonmelanoma skin cancer management. Dermatol Surg 2006; 32: 1266–1271.
Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH . A single type of progenitor cell maintains normal epidermis. Nature 2007; 446: 185–189.
Levy V, Lindon C, Harfe BD, Morgan BA . Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 2005; 9: 855–861.
Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 2010; 327: 1385–1389.
Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol 2010; 12: 299–305.
Gailani MR, Stahle-Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet 1996; 14: 78–81.
Lauth M, Toftgard R . The Hedgehog pathway as a drug target in cancer therapy. Curr Opin Investig Drugs 2007; 8: 457–461.
Oro AE, Higgins KM, Hu Z, Bonifas JM, Epstein Jr EH, Scott MP . Basal cell carcinomas in mice overexpressing sonic hedgehog. Science 1997; 276: 817–821.
Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998; 391: 90–92.
Teglund S, Toftgard R . Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta 2010; 1805: 181–208.
Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H et al. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol 2009; 130: 124–134.
Yi R, Poy MN, Stoffel M, Fuchs E . A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 2008; 452: 225–229.
Adolphe C, Hetherington R, Ellis T, Wainwright B . Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res 2006; 66: 2081–2088.
Hutchin ME, Kariapper MS, Grachtchouk M, Wang A, Wei L, Cummings D et al. Sustained Hedgehog signaling is required for basal cell carcinoma proliferation and survival: conditional skin tumorigenesis recapitulates the hair growth cycle. Genes Dev 2005; 19: 214–223.
Kasper M, Jaks V, Are A, Bergstrom A, Schwager A, Barker N et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci USA 2011; 108: 4099–4104.
Laner-Plamberger S, Kaser A, Paulischta M, Hauser-Kronberger C, Eichberger T, Frischauf AM . Cooperation between GLI and JUN enhances transcription of JUN and selected GLI target genes. Oncogene 2009; 28: 1639–1651.
Schnidar H, Eberl M, Klingler S, Mangelberger D, Kasper M, Hauser-Kronberger C et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res 2009; 69: 1284–1292.
Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M et al. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS ONE 2007; 2: e610.
Eferl R, Wagner EF . AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003; 3: 859–868.
Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA 1999; 96: 9827–9832.
Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M . Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J 2000; 19: 2056–2068.
Mainiero F, Murgia C, Wary KK, Curatola AM, Pepe A, Blumemberg M et al. The coupling of alpha6beta4 integrin to Ras-MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J 1997; 16: 2365–2375.
Carleton M, Cleary MA, Linsley PS . MicroRNAs and cell cycle regulation. Cell Cycle 2007; 6: 2127–2132.
Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 2006; 38: 356–362.
Wei T, Orfanidis K, Xu N, Janson PC, Ståhle M, Pivarcsi A et al. The expression of microRNA-203 during human skin morphogenesis. Exper Dermatol 2010; 19 (9): 854–856.
Zenz R, Wagner EF . Jun signalling in the epidermis: from developmental defects to psoriasis and skin tumors. Int J Biochem Cell Biol 2006; 38: 1043–1049.
Herber B, Truss M, Beato M, Muller R . Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 1994; 9: 1295–1304.
Sharpe GR, Fisher C, Redfern CP . Changes in oncogene mRNA expression during human keratinocyte differentiation. Arch Dermatol Res 1994; 286: 476–480.
Pivarcsi A, Muller A, Hippe A, Rieker J, van Lierop A, Steinhoff M et al. Tumor immune escape by the loss of homeostatic chemokine expression. Proc Natl Acad Sci USA 2007; 104: 19055–19060.
Ikebe D, Wang B, Suzuki H, Kato M . Suppression of keratinocyte stratification by a dominant negative JunB mutant without blocking cell proliferation. Genes Cells 2007; 12: 197–207.
Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 2008; 40: 1291–1299.
Acknowledgements
We thank Anna-Lena Kastman and Ulrica Westermark for excellent technical support. This work was supported by the Cancerfonden (09 0788), the Swedish Research Council (K2008-67X-20778-01-3 and DNR: 2011-2621), Medical Research Council (K2008-74x-07133-21A), Karolinska Institutet, Welander Finsens Foundations, Tore Nilsons Foundation, Konsul ThC Bergh Foundations and the Stockholm County Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogenesis website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sonkoly, E., Lovén, J., Xu, N. et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis 1, e3 (2012). https://doi.org/10.1038/oncsis.2012.3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2012.3
Keywords
This article is cited by
-
Identification and functional validation of SRC and RAPGEF1 as new direct targets of miR-203, involved in regulation of epidermal homeostasis
Scientific Reports (2023)
-
Investigating melanogenesis-related microRNAs as disease biomarkers in vitiligo
Scientific Reports (2022)
-
Integrative analysis of mRNA-miRNA-TFs reveals the key regulatory connections involved in basal cell carcinoma
Archives of Dermatological Research (2020)
-
MicroRNA-203 suppresses proliferation in liver cancer associated with PIK3CA, p38 MAPK, c-Jun, and GSK3 signaling
Molecular and Cellular Biochemistry (2018)
-
The Role of Exosomes in Pancreatic Cancer Microenvironment
Bulletin of Mathematical Biology (2018)