Abstract
Tumor surveillance of natural killer (NK) cells is mediated by the cytotoxicity receptor natural-killer group 2 member D (NKG2D). Ligands for NKG2D are generally not expressed on healthy cells, but induced on the surface of malignant cells. To date, NKG2D ligand (NKG2D-L) induction was mainly described to depend on the activation of the DNA damage response, although the molecular mechanisms that regulate NKG2D-L expression remain largely unknown. Here, we show that the acetyltransferases CBP (CREB-binding protein) and p300 play a crucial role in the regulation of NKG2D-L on tumor cells. Loss of CBP/p300 decreased the basal cell surface expression of human ligands and reduced the upregulation of MICA/B and ULBP2 in response to histone deacetylase inhibitors or DNA damage. Furthermore, CBP/P300 deficiency abrogated the sensitivity of stressed cells to NK cell-mediated killing. CBP/p300 were also identified as major regulators of mouse NKG2D ligand RAE-1 in vitro and in vivo using the Eμ-Myc lymphoma model. Mechanistically, we observed an enhanced activation of the CBP/p300 binding transcription factor CREB (cAMP response element-binding protein) correlating to the NKG2D-L upregulation. Moreover, increased binding of CREB and CBP/p300 to NKG2D-L promoters and elevated histone acetylation were detectable. This study provides strong evidence for a major role of CBP and p300 in orchestrating NKG2D-L induction and consequently immunosurveillance of tumors in mice and humans. These findings might help to develop novel immunotherapeutic approaches against cancer.
Similar content being viewed by others
Introduction
One of the major natural killer (NK) cell receptors involved in recognition and killing of tumor cells is the cytotoxic receptor, natural-killer group 2 member D (NKG2D).1 NKG2D is expressed on NK cells and CD8+ T cells, some γδ T cells and possibly also some CD4+ T cells and is known as a sensor for damaged or dangerous cells. In humans, NKG2D is engaged by several ligands, namely major histocompatibility complex (MHC) class I polypeptide-related sequence A and B (MICA and MICB) and the UL16-binding proteins 1–6 (ULBP1–6).2 Mouse ligands binding to NKG2D are the GPI-linked retinoic acid early inducible-1 (RAE-1) proteins,3 the transmembrane protein murine UL16-binding protein-like transcript 1 (MULT1)4 and the histocompatibility 60 (H60) family.5
In vivo evidence for the significance of NKG2D in eradication of cancer has been observed in several mouse models of spontaneous malignancies.6 More recently, transplantation experiments and the λ-Myc transgenic lymphoma model were used to show that NKG2D engagement is critical for immunosurveillance of lymphomas and that selection for NKG2D ligand (NKG2D-L) loss mutants provides a mechanism of tumor escape.7
Tumor cells develop mechanisms to escape from innate immune surveillance and these strategies include shedding of NKD2D-Ls from target cells to inhibit NK cell activity as demonstrated in many studies for different tumor entities.8
Although NKG2D-Ls are not expressed on healthy cells, they are upregulated within different disease contexts—including infection, transformation, extensive proliferation, wound repair and inflammatory diseases. The molecular pathways directing their inducible expression are still not defined and depend on transcriptional, translational and post-translational regulation.2, 9 The DNA damage response (DDR) kinases ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia and RAD3 related) are involved in the NKG2D-L upregulation in response to DNA damage by tumor cells, initially demonstrated in response to radiation and chemotherapy.10, 11 Expression of ligands in response to many small molecules such as an inhibitor for HSP90(ref. 12) or IAP (inhibitor of apoptosis) inhibitors was attributed to their ability to activate the DDR.13 Nevertheless, downstream signaling remains elusive. Given the existence of different NKG2D-Ls and their induced expression, a complex, heterogeneous and context-dependent regulation seems likely. Not surprisingly, a contribution of diverse transcription factors including heat shock pathway, E2F, family of Sp transcription factors, AP-1, AP-2a, p53 and nuclear factor (NF)-κB was reported.2, 9 However, their impact varied depending on the cell line or the model system used, as described for example for the p53-dependent NKG2D-L induction.14, 15, 16 Here we show that the major acetyltransferases CBP and p300 have a robust, mandatory and general impact on the upregulation of NKG2D-Ls MICA/B and ULBP2 in humans and RAE-1 in mice.
Results
HDACis induced NKG2D-L expression independently of the DDR
Initially, several cell lines were screened for MICA/B induction upon diverse stimuli to induce DNA damage and with inhibitors of histone deacetylases (HDACis) to establish an experimental setting with a strong and reproducible upregulation in a noncell type-specific manner to be used for future experiments. Of note, none of the tested DNA-damaging agents induced a robust upregulation of MICA/B (Figure 1a). In contrast, the HDACis trichostatin A (Figure 1a) and LBH589 (not shown) induced a significant NKG2D-L upregulation in virtually all tested cell lines. Moreover, a panel of HDAC class-specific inhibitors with specificity for the different subsets of histone deacetylases induced the MICA/B surface expression (Figure 1b).
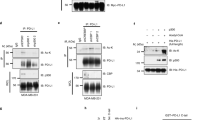
HDACis were potent inducers of the NKG2D-Ls MICA/B in humans. (a) Indicated cell lines were incubated with a panel of diverse DNA-damaging agents (gemcitabine (Gem), 2 μM; cytarabine (Ara-C), 10 μM; aphidicolin (Aph), 20 μM; bleomycin (Bleo), 30 μg/ml; and cisplatin (Cis), 10 μg/ml) and the HDACi TSA (trichostatin A, 250 nM) for 16 h. Flow cytometry (dead cells were excluded for analysis) was performed for surface expression of MICA/B. (b) MDA-231 cells were treated with a panel of different HDACis (TSA, 250 nM; CAY-10683, 5 μM; MS-275, 5 μM; scriptaid (SCR), 5 μM; SIRT1 inhibitor, 50 μM; and SIRT2 inhibitor, 50 μM) for 16 h and analyzed by FACS. Mean±s.d. of two or more independent experiments is indicated.
For most of the subsequent experiments, we used the HDACi LBH589 (panobinostat) as it upregulated NKG2D-L at lower concentration than most other HDCAis (see Figure 2d).
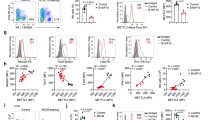
HDACi-induced NKG2D ligand regulation did not involve the DDR. (a) HEK-293 cells were treated with 100 nM LBH589 for 16 h, washed and incubated with primary human NK cells of healthy donors in indicated ratios for 3 h. Mean and s.e. of four independent experiments. (b) FACS analysis of HEK-293 cells after 1 h of preincubation with 2 μM actinomycin D or 10 μM cycloheximide, and treatment with LBH589 (100 nM) for 6 h. (c) Real-time PCR for HEK-293 cells treated with 10 μM Ara-C or 100 nM LBH589 for the indicated periods relative to GAPDH. (d) HEK-293 cells were treated with the ATM/ATR inhibitor CGK733 (5 μM) together with the damage inducer Ara-C (10 μM) or the HDACis LBH589 (100 nM) and trichostatin A (TSA; 250 nM) for 16 h to analyze the surface expression of MICA/B, ULBP1, ULBP2 and ULBP3. (e) MDA-231 and 911 cells were incubated with ATM/ATR inhibitor CGK733 (5 μM) or ATM inhibitor KU55933 (10 μM) and TSA (250 nM) for 16 h and subsequently studied for MICA/B expression by FACS. (f) Intracellular FACS staining of HEK-293 cells for DNA damage marker phospho-γH2AX (S139 phosphorylated) after incubation with 10 μM Ara-C or 100 nM LBH589 for 1 h.
To test the functional significance of HDACi-induced NKG2D-L expression, we analyzed the sensitivity of HDACi-treated cells to NK cell-mediated lysis. LBH589-treated HEK-293 cells were significantly more lysed by NK cells compared with control-treated HEK-293 cells (Figure 2a). This suggests that the HDACi-induced NKG2D-L upregulation is functionally important. Next, we demonstrated that treatment of tumor cells with HDACi also upregulated NKG2D-L transcript levels. Pretreatment of cells with the transcription inhibitor actinomycin D or the translation inhibitor cycloheximide completely abrogated the LBH589-mediated induction of MICA/B on the cell surface (Figure 2b) implying that their upregulation depends on de novo transcription. In line, NKG2D-L mRNA upregulation was observed as early as after 1 h of HDACi treatment (Figure 2c).
Although MICA, MICB and ULBP2 were significantly upregulated, the induction of ULBP1 and -3 by HDACis was comparable to the effects of the DNA damage inducer Ara-C (Figure 2d).
HDACis were previously found to regulate NKG2D-L via the DDR.10, 17, 18 Unexpectedly, ATM/ATR inhibitors only partially blocked the LBH589- or trichostatin A-mediated upregulation of MICA/B and ULBP2, whereas they inhibited the induction of NKG2D-L by the DNA-damaging agent Ara-C (Figures 2d and e). Accordingly, no phosphorylation of the DDR markers CHK1 (not shown) and γH2AX was observed after treatment of cells with different HDACis (Figure 2f). In summary, the data suggest that HDACis are efficient inducers of NKG2D-L expression. NKG2D-L upregulation happened on transcriptional level, was of biological relevance and seemed to be independent of the DDR.
Induction of NKG2D ligands by HDACis was dependent on the acetyltransferases CBP/p300
The viral p300-binding oncogene E1A was shown to regulate NKG2D-L expression19 and we therefore analyzed the role of acetyltransferases CBP/p300 in HDACi-induced NKG2D-L expression. CBP (also known as CREB-binding protein or CREBBP) and p300 (also known as EP300 or E1A binding protein p300) are two closely related transcriptional co-activating proteins with acetytransferase activity.
To this end, the impact of acetyltransferase inhibitors was anaylzed. Anacardic acid (ANAC) is not specific for CBP/p300 and inhibits a broad spectrum of acetyltransferases; C646 is specific for CBP/p300 and KIXi inhibits the binding of CBP/p300 to the transcription factor CREB.20 Intracellular staining of HEK-293 cells revealed enhanced binding of an anti-acetyl-lysine antibody upon HDACi treatment, reflecting a general increase in acetylation as expected. The general acetylation was reduced by pretreatment of cells with ANAC or with the CBP/p300 inhibitor C646 (Figure 3a). Of note, inhibition of acetylation by ANAC or C646 was sufficient to significantly impair MICA/B upregulation upon HDACi treatment in HEK-293 cells (Figures 3b and c). Notably, C646 also blocked Ara-C-induced NKG2D-L induction (Figure 3b). Inhibition of CBP/p300 abolished the up-regulation of NKG2D-L transcripts by HDACis, suggesting that the acetyltransferases CBP/p300 regulate the transcription of NKG2D-L (Figure 3d). To investigate the impact of CBP/p300-mediated acetylation on NKG2D-L upregulation on tumor cells, we treated a panel of tumor cell lines from different origins with acetyltransferase inhibitors (Figure 4). A significant diminished expression of MICA/B and ULBP2 was observed in all cell lines tested, thus confirming that CBP/p300 contributes to the upregulation of NKGD-L on tumor cells. Killing assays using L428 cells as targets revealed that the HDACi-dependent upregulation of NKG2D-Ls rendered target cells more susceptible to NKG2D-dependent killing, as expected (Supplementary Information S1.
Induction of NKG2D-Ls by HDACis was dependent on the acetyltransferases CBP/p300. (a) Intracellular FACS analysis of HEK-293 cells stained for acetylated lysine residues. Cells were preincubated with or without 75 μM of the histone acetyltransferase inhibitor (HATi) anacardic acid (ANAC) or 10 μM of the p300 (and CBP) inhibitor C646 for 3 h and subsequently treated with 100 nM LBH589 for 1 h. (b) Flow cytometric analysis of HEK-293 cells treated with or without 8 μM C646 and 10 μM Ara-C or 250 nM trichostatin A (TSA) for 16 h. (c) HEK-293 cells were incubated with 50 μM ANAC, 8 μM C646 or 10 μM CBP-CREB interaction inhibitor (KIXi) and treated with 100 nM LBH589 (LBH). (d) Real-time PCR of HEK-293 cells preincubated with or without 10 μM C646 for 3 h and treated with 100 nM LBH589 for 1 h. MICA, MICB and ULBP2 mRNA expression levels were analyzed relative to GAPDH.
CBP/p300-dependent NKG2D-L induction by HDACi was observed in a panel of tumor cell lines from different origins. Indicated cell lines originating from (a) hematological tumors and (b) solid tumors were incubated with 50 μM ANAC, 8 μM C646, or 10 μM CBP-CREB interaction inhibitor (KIXi) and treated with 100 nM LBH589 for 16 h followed by flow cytometric analysis using the indicated NKG2DL-specific antibodies. Mean and standard error of three independent experiments.
To formally prove the role of CBP/p300 in NKG2D-L induction, we used CBP/p300-deficient HEK-293 cells. A CRISPR/Cas9 nuclease (dKOnuc) and a nickase (dKOnic) approach was used to generate two independent CBP/p300 double-knockout clones clones (mutation analysis, see Supplementary Information S2). CBP/p300 expression was significantly reduced at the protein and transcript levels (Figure 5a), although not completely abolished. In line, intracellular staining showed decreased general acetylation levels in CBP/p300-deficient cells (Figure 5b).
CBP/p300-deficient cells revealed a diminished NKG2D-L upregulation in response to HDACi and DNA damage. (a–g) HEK-293 CBP/p300 double-knockout (dKO) cells were generated using the CRISPR/Cas9 nuclease (dKOnuc) or CRISPR/Cas9 nickase (dKOnic) approach. (a, left panel) Western blot of CBP/p300 dKO or WT HEK-293 cells using an anti-CBP antibody (C-terminal, clone A22) detecting both CBP and p300. GAPDH was used as loading control. (a, right panel) Real-time PCR for CBP and p300 mRNA expression levels relative to GAPDH. (b) Basal expression levels of MICA/B measured by flow cytometry. (c) Cells were treated with 100 nM LBH589 for indicated time periods and MICA/B expression was analyzed by FACS. (d, e) Flow cytometric analysis of MICA/B (left panel) and ULBP2 (right panel) of CBP/p300 dKO or WT HEK-293 treated with 100 nM LBH589 or 10 μM Ara-C for 16 h. (f) Intracellular FACS for acetylated lysine after 1 h of treatment with 100 nM LBH589. (g) FACS-based NK cell killing assay. CBP/p300 dKO or WT HEK-293 cells were treated with 100 nM LBH589 for 16 h, washed and incubated with primary human NK cells in indicated ratios for 3 h. (g, left panel) One representative experiment and (g, right panel) mean and s.e. of four independent experiments (ratio 5:1).
Loss of CBP/p300 decreased the basal cell surface expression of MICA/B and reduced the upregulation of MICA/B and ULBP2 in response to HDACis and Ara-C compared with control HEK-293 cells (Figures 5c–f) suggesting that CBP/p300 play an important role in the constitutive and inducible expression of NKG2D-L. Furthermore, CBP/p300 deficiency abrogated the increased sensitivity of LBH589-treated cells to NK cell-mediated lysis (Figure 5g).
Of note, the induction of MICA/B and ULBP2 in response to HDACis was independent of NF-κB and p53 acetylation, not influenced by NF-κB or p53 inhibitors and observed in syngeneic wild-type p53 and p53-deficient cells (Supplementary Information S3). Conclusively, these results reveal the important role of CBP/p300 in the regulation of NKG2D-L in human cancer cells.
HDACi treatment induced enhanced binding of acetylated histone H3, CBP/p300 and CREB to promoter regions of human NKG2D-L genes
Next, we used a phospho-kinase profiler array to identify potential CBP/p300 targets involved in NKG2D-L upregulation (Figure 6a). Several of the analyzed kinases did not show enhanced phosphorylation after treatment with LBH589 (for instance, HSP27). Some others were induced by LBH589 but induction could not be blocked by the CBP/p300 inhibitor C646 (for example, β-catenin, Akt and extracellular signal-regulated protein kinases 1 and 2 (ERK1/2)). Most interestingly, the array revealed that CREB phosphorylation increased upon incubation with LBH589 that was abolished by C646 treatment. Accordingly, enhanced CREB binding to NKG2D-L promoter regions was found by chromatin immunoprecipitation (Figure 5b). This was in line with the observation that a CBP–CREB interaction inhibitor (KIXi) significantly blocked LBH589-induced MICA/B upregulation on the cell surface (Figure 2c). In addition, acetylation of histone H3 at the MICA, MICB and ULBP2 promoters was significantly augmented in response to LBH589 (Figure 6b).
HDAC inhibition induced enhanced binding of acetylated histone H3, CBP/p300 and CREB to NKG2D-ligand promoters. (a) Phospho-kinase profiler array of HEK-293 cells pretreated with or without 10 μM C646 for 3 h and incubated with 100 nM LBH589 for one additional hour, followed by lysis of the cells. Detection of phosphorylated kinases was performed with a digital ECL imager. (b) Chromatin immunoprecipitation (ChIP) of HEK-293 cells treated with 100 nM LBH589 for 3 h. Pull down was performed with antibodies against acetylated histone H3, CBP (also binding to p300) and CREB. Precipitated promoter sequences were detected by real-time PCR and calculation was implemented using the % input method. Values were normalized to RPL30.
STAT (signal transducer and activator of transcription) signaling is one of the nonhistone targets of CBP/p300. Of note, STAT5a/b and STAT6 phosphorylation increased upon LBH589 treatment that was partially blocked when the cells were preincubated with the CBP/p300 inhibitor C646 (Figure 5a). Although it was not possible to confirm STAT activation by conventional western blot or intracellular flow cytometric analysis in our setting (not shown), it is tempting to speculate that CBP/p300 act at least in part via STAT signaling to induce NKG2D-L expression.
To address the role of CBP/p300 in NKG2D-L expression in cancer cells in vivo, we bred mice that specifically lacked CREBBP(CBP) and EP300(p300) in CD19+ B cells21 with the Eμ-Myc lymphoma strain.22, 23 B-cell lymphomas in Eμ-Myc mice express NKG2D-L and NKG2D-deficient Eμ-Myc mice show an accelerated development of B-cell lymphomas, implicating a role for NKG2D in tumor surveillance.6 Genotyping of the littermates showed that either CBP or p300 was deleted, but never both genes (Figure 7a), indicating that the activity of at least one of the acetyltransferases is indispensable for B-cell development and/or survival. As soon as first signs of tumors were detectable (male and female, age of mice was between 86 and 159 days) tumor cells were isolated from lymph nodes, spleen and peripheral blood to analyze NKG2D-L expression. Strikingly, surface expression of RAE-1 was significantly reduced in CBP/p300-deficient Eμ-Myc tumor cells (Bnull) compared with their CBP/p300-proficient counterparts (ctrl) (Figure 7b). The diminished RAE-1 surface expression correlated with reduced RAE-1 transcript levels (Figure 7c). Interestingly, the expression of MULT1 remained unaffected, indicating that MULT1 and RAE-1 are regulated independently, and this might reflect different biological functions of these ligands.24 Finally, these data are consistent with in vitro data showing that CBP/p300 inhibition blocked RAE-1 induction on mouse MCA-205 cells, whereas MULT1 expression remained stable (Figure 7d). In summary, we identified CBP/p300 as a major regulator of mouse NKG2D-L RAE-1 in vitro and in vivo.
CBP/p300 were crucial for the expression of RAE-1 in vivo. (a) Eμ-Myc CBP/P300 littermates show deletion of either CBP or p300. Genomic DNA extracted from tumor cells of terminally ill Eμ-Myc CBP/P300 double mutants (Bnull) was subjected to PCR using specific primers to detect recombined (Δflox) or non-recombined (flox) genes. Representative examples are shown. (b) Flow cytometric analysis of tumor cells from lymph nodes to detect MULT1 and RAE-1 (right panel) and of tumor cells from lymph nodes (tumor), spleen or peripheral blood (PB) (left panel) isolated from Eμ-Myc mice (ctrl) or Eμ-Myc with CBP/p300-deficient B cells (Bnull). (c) Real-time PCR to detect RAE-1 transcripts expressed in tumor cells from Eμ-Myc mice (ctrl) or Eμ-Myc with CBP/p300-deficient B cells (Bnull). RAE-1 mRNA expression was analyzed relative to HPRT. (d) Flow cytometric analysis of MCA-205 cells that were preincubated with 8 μM C646 and treated with 5 nM LBH589 for 16 h followed by detection of MULT1 and RAE-1 surface expression using flow cytometry.
No differences in lymphoma onset, tumor load or survival were observed between control Eμ-Myc mice and Eμ-Myc mice with CBP/p300-deficient B cells (data not shown). It is possible that MULT1 expression counterbalances the RAE-1 deficiency in the model used. However, differences between the two groups might be masked by the heterogeneity of tumor onset observed in this model (onset of tumor was between 3 and 5 months).
Discussion
NKG2D is one of the best-characterized activating NK cell receptors that promotes NK cell-mediated lysis of tumor cells and plays a major role in tumor surveillance. The identification of key molecules that regulate the inducible expression of ligands for NKG2D on target cells is pivotal to fully decipher NK cell-mediated defense and crucial to develop immunotherapeutic approaches that aim to sensitize tumor cells for the NKG2D-dependent tumor clearance. Here, we provide evidence for a critical role of CBP/p300 in the transcriptional regulation of ligands engaging NKG2D. Our conclusion is based on the findings that HDACi treatment resulted in a robust and strong upregulation of NKG2D-L even compared with the treatment with DNA-damaging agents in various human and murine cell lines (1), and this upregulation was blocked upon chemical inhibition or genetic ablation of acetyltransferases CBP/p300 in vitro and in vivo (2), causing a reduced NK cell-mediated killing (3). Finally, we observed elevated histone acetylation and enhanced CBP/p300 and CREB binding at NKG2D-L promoter regions. These data suggest that CBP/p300 mediate transcriptional activation of NKG2D ligands by induction of a more open chromatin state at NKG2D-L genes, by mediating CREB binding to NKG2D promoters and possibly via acetylating unknown transcription factors thereby enhancing their activity. The critical role of CBP/p300 was shown for human NKG2D-Ls MICA/B and ULBP2 and for the mouse ligand RAE-1.
These data suggest that MICA and MICB, together with ULBP2 and RAE-1, are regulated in another way than ULBP1, ULBP3 and MULT1. All of them with the exception of MULT1 could be induced by HDACis, but to dissimilar amplitudes. A distinct regulation pathway of MULT1 might reflect its special function among NKG2D-L. Although it is common sense that secreted ligands for NK cell receptors generally impair NK cell function, the shed soluble form of MULT1 actually enhanced NK cell activity and caused tumor rejection.24 This is conclusive with our own data indicating an activating role of secreted BAG6, a ligand for the activating receptor NKp30, as long as it is bound to exosomes.25, 26, 27, 28. This provides clear evidence to revisit the paradigm that secreted ligands for NK cell receptors are always inhibitory and counteract immunosurveillance.
To date, HDACi-mediated NKG2D-L induction was mainly described to depend on activation of the DDR and ATM/ATR.29 However, inhibiting ATM/ATR using CGK733 or ATM using KU55933 failed to block HDACi-induced NKG2D-Ls (Figures 2d–f), suggesting that HDACi-mediated NKG2D-L upregulation was independent of the DDR.
One putative downstream target of CBP/p300 that might be involved in NKG2D-L upregulation was NF-κB. NF-κB had already been proposed to play a role in NKG2D-L induction in T cells30, 31 and endothelial cells.32 Furthermore, several groups claimed that HDACis activate NF-κB.33, 34, 35 In line with others,36 this study clearly argues against a general role of NF-κB in the regulation of NKG2D-L as it was not possible to detect HDACi-mediated NF-κB activation (Supplementary Information S3). Recent findings suggest a role for p53 in the regulation of the NKG2D-L ULBP2.14, 37, 16 Similar to NF-κB, a role for p53 in HDACi-dependent regulation could not be confirmed in this study (Supplementary Information S3). This is in line with the results of others who also claim that p53 is not involved in NKG2D-L upregulation.10, 38 Interestingly, a role for p53 seems actually restricted to selected cell lines,16 indicating that the impact of p53 on NKG2D-L expression depends on cell specific conditions or the environment. Specific transcription factors that upregulate ligands for NKG2D recruited by the acetyltransferases CBP/p300 remain to be identified.
A role for CBP/p300 as tumor suppressor genes has been already suggested.39 Moreover, somatic mutations were reported for both or either of these genes in epithelial cancers,40 B-cell non-Hodgkin’s lymphoma,41 endometrial tumors42 and acute lymphocytic lymphoma.43 Although we clearly showed that RAE-1 expression was impaired in CBP/p300-deficient Eμ-Myc cells, we did not observe any impact on tumor onset or course. The Eμ-Myc transgenic mice express the myc oncogene constitutively under the control of the IgH enhancer and develop spontaneous tumors of the B-cell lineage within the first 5 month of life.22, 23 As it was shown that NKG2D-mediated surveillance is operative for these lymphomas,6 we speculate that MULT1 expression might compensate the RAE-1 deficiency in the model used. However, we cannot exclude that differences are masked by the enormous heterogeneity of tumor onset and progression observed in this model.
HDACs are an established and validated target for the treatment of cancer and some HDACis have already been approved by the US-FDA (US Food and Drug Administration).44 Parts of the clinical benefit of HDAC inhibition were already attributed to their impact on the immune cells. However, HDAC inhibition features immune activating as well as immune inhibiting effects.45 Dual therapy with HDACis together with immunotherapy have shown promising results in preclinical studies.46
Here, we provide evidence for a major role of CBP/p300 in the regulation of ligands for the activating NK cell receptor NKG2D. HDACi-mediated CBP/p300-dependent NKG2D-L induction was able to enhance in vitro killing by NK cells, emphasizing the biological significance of these results. Transcriptional activation of NKG2D-L happened via interplay of enhanced histone acetylation resulting in a more accessible chromatin state, together with enhanced binding of CBP/p300 as well as CREB to the promoter regions of NKG2D-L genes. These findings shed more light onto the complex regulation of ligands for activating NK cell receptors that are important mediators of immune defense against cancer. Further research in this field will not only help to understand the basic biology of NK cell-mediated tumor surveillance but will also pave the way toward exploiting this mechanism to facilitate the patient’s immune system to combat cancer.
Materials and methods
Cell lines
Cell lines (see list in Supplementary Information S5) were maintained in RPMI-1640 GlutaMAX (Invitrogen, Darmstadt, Germany) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution at 37 °C and 5% CO2. Regular mycoplasma testing was performed by PCR and only mycoplasma-negative cells were used in experiments. It was avoided that cells overgrew before splitting because this might influence NKG2D-L expression.
Before treatment, adherent cells were seeded to reach 70% confluence on the day of harvesting. For suspension cells, 5 × 105 cells per ml were used. Untreated controls were incubated with equivalent volume of the vehicle dimethyl sulfoxide.
Inhibitors/cytostatic agents
Reagents were purchased from SCBT (Heidelberg, Germany) if not stated otherwise. The reagents and the concentrations used were actinomycin D (2 μM), cycloheximide (10 μM), the histone acetyltransferase inhibitor (ANAC (50 μM for 16 h of incubations or 75 μM for shorter incubations) and the HDACis CAY-10683 (5 μM), MS-275 (5 μM), Panobinostat (LBH589) (100 nM for human cells or 5 nM for mouse cells), scriptaid (5 μM), SIRT1 inhibitor IV (50 μM), SIRT2 inhibitor (50 μM) and trichostatin A (250 nM). DNA damage was induced with Ara-C (cytarabine) (10 μM for human cells or 500 nM for mouse cells). The CBP/p300 inhibitor C646 (8 μM for 16 h of incubations or 10 μM for shorter incubations in serum-free medium), the CBP–CREB interaction inhibitor (‘KIXi’) (10 μM) (Calbiochem, Merck Millipore, Darmstadt, Germany) and the ATM/ATR inhibitor GK733 ATM/ATRi (5 μM) were used. All agents were dissolved in dimethyl sulfoxide.
Flow cytometry
Fluorescence-activated cell sorting (FACS) was performed on a FACSCalibur (Becton Dickinson, Heidelberg, Germany) or Gallios (Beckman Coulter, Krefeld, Germany). 7-Aminoactinomycin D (7-AAD; BioLegend, Koblenz, Germany) was added before measurement and only negative cells were included in analysis. The following specific antibodies were used: ac-Lysine (9441, CST, Leiden, Netherlands); MICA (AMO1, BamOmaB); MICA/B (6D4, BioLegend); MICB (BMO2, BamOmaB, Gräfelfing, Germany); MULT-1 (237104, R&D); p-γH2AX (S139, 9718, CST); Rae-1 (R&D, Wiesbaden-Nordenstadt, Germany); ULBP1 (AUMO2, BamOmaB); ULBP2 (BUMO1, BamOmaB); ULBP2 (BAF1298, R&D); ULBP3 (CUMO3 BamOmaB) and isotype controls purchased from BioLegend.
Western blot
Whole-cell lysates were prepared with cell lysis buffer from CST (9803). Anti-GAPDH (horseradish peroxidase conjugated, 8884, CST) and A-22 (SCBT) were used to detect glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and CBP/p300, respectively. Proteins were detected with ECL (Thermo Scientific, Darmstadt, Germany).
Quantitative reverse transcription–PCR and genotyping PCR
RNA was extracted from cell lines using the M&N NuceloSpin (Machery-Nagel, Düren, Germany) RNA kit and 1 μg RNA was used for complementary DNA synthesis (RevertAid First Strand cDNA kit from Thermo Scientific). Real-time PCR was performed using Sigma (Hamburg, Germany) SYBR Green JumpStart in the Life Technologies (Thermo Scientific) 7500 real-time PCR system. Initial heat inactivation was performed for 15 min at 95 °C. Then, 40 cycles of 15 s at 94 °C, 30 s at 56 °C and 30 s at 72 °C were followed by melting curve analysis. See list of primers in Supplementary Information S4A.
Mice were genotyped by semiquantitative PCR using CBP-specific primers (5′-CTCTACATCCTAAGTGCTAGG-3′ and 5′-CAGTAGATGCTAGAGAAAGCC-3′, producing a 380-bp wild-type band, a 530-bp p300 flox band and a 950-kb p300 Δflox band) and using p300-specific primers (5′-GGGGAAATTTTGGCTGGCAAG-3′ and 5′-CTGCTCTACCTAAATTCCCAG-3′, producing 1250, 1100 and 970 bp fragments for the wild-type, floxed and Δflox alleles, respectively).
PCR was run in a C1000 Touch Thermal Cycler (Bio-Rad, München, Germany) with 5 min at 94 °C, 36 × (10 s at 94 °C, 1 min at 55 °C and 2 min at 68 °C) and 10 min at 72 °C.
CRISPR/Cas9-mediated knockout
Knockout of CBP and p300 was performed using the Zhang Lab’s reagents and protocols (Massachusetts Institute of Technology, Cambridge, MA, USA), for details see Supplementary Information S2.
NK cell killing assay
Peripheral blood mononuclear cells were isolated from buffy coats of healthy blood donors that were provided by the blood bank of the University Hospital Cologne (approval by the ethics committee: 08-275). Primary NK cells were isolated using the human NK cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). HEK-293 target cells were stained with Life Technologies cell membrane dye DiR (800 ng/ml for 20 min at 37 °C in serum-free medium followed by two washing steps with serum-containing medium) before coincubation with NK effector cells in different ratios. Dead target cells were then identified by 7-AAD staining (BioLegend) and relative cytotoxicity was calculated by 100 × [(% dead target cells in sample−% spontaneous dead target cells)/(100−% spontaneous dead target cells)].
Phospho-kinase profiler array
The human phospho-kinase profiler array (R&D Systems) was performed according to the manufacturer’s protocol. Read-out was conducted using a digital cheminoluminescence imager. Data were analyzed using ImageJ (from Wayne Rasband, National Institute of Health (NIH), USA).
Chromatin immunoprecipitation
Primers (see Supplementary Information S4B) were designed to amplify promoter regions after prior in silico analysis for probable transcription factor binding sites (http://www.cbrc.jp/cbrc-software). Chromatin immunoprecipitation was conducted using the SimpleChIP Enzymatic Chromatin IP Kit from CST (9003) according to the manufacturer’s protocol. Real-time PCR (primers see Supplementary Information S4B) was executed with purified DNA in the Life Technologies 7500 Real-time PCR system. Initial heat inactivation was performed for 15 min at 95 °C. Then, 40 cycles of 15 s at 94 °C, 30 s at 56 °C and 30 s at 72 °C were followed by melting curve analysis.
Animals
Eμ-Myc transgenic mice22, 23 were crossed with CBPfl/fl p300fl/fl CD19Cretg/wt (both C57BL/6)21 to obtain the genotype CBPfl/fl;p300fl/fl;CD19Cretg/wt;EμMyctg/wt. We have the permission for breeding (Zuchtrahmenantrag: 84-02.04.2014.A146; Anzeigen: 84-02.05.201 З.071 and 84-02.05.201 З.070) and the animal experiments were approved by the Landesamt für Umwelt und Verbraucherschutz Nordrhein-Westfalen (Aktenzeichen 84-02.04.2012.A.216). For animal studies randomization or blinding was not applicable (no treatment groups).
Statistics
GraphPad Prism 6 was used for depicting bar charts of means with s.e.m. and for statistics (*P<0.05, **P<0.01, ***P<0.001). Student’s t-test was used for statistical analyses of normally distributed measurements and Wilcoxon signed-rank test was used if values were not normally distributed. The variance between the groups that were statistically compared was similar.
References
Bauer S . Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible mica. Science 1999; 285: 727–729.
Lanier LL . NKG2D receptor and its ligands in host defense. Cancer Immunol Res 2015; 3: 575–582.
Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 2000; 12: 721–727.
Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM . Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J Immunol 2002; 169: 4079–4083.
Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH . Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol 2000; 1: 119–126.
Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008; 28: 571–580.
Belting L, Homberg N, Przewoznik M, Brenner C, Riedel T, Flatley A et al. Critical role of the NKG2D receptor for NK cell-mediated control and immune escape of B-cell lymphoma. Eur J Immunol 2015; 45: 2593–2601.
Champsaur M, Lanier LL . Effect of NKG2D ligand expression on host immune responses. Immunol Rev 2010; 235: 267–285.
Raulet DH, Gasser S, Gowen BG, Deng W, Jung H . Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol 2013; 31: 413–441.
Gasser S, Orsulic S, Brown EJ, Raulet DH . The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 2005; 436: 1186–1190.
Lam AR, Le Bert N, Ho SS, Shen YJ, Tang ML, Xiong GM et al. RAE1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res 2014; 74: 2193–2203.
Boll B, Eltaib F, Reiners KS, von Tresckow B, Tawadros S, Simhadri VR et al. Heat shock protein 90 inhibitor BIIB021 (CNF2024) depletes NF-kappaB and sensitizes Hodgkin's lymphoma cells for natural killer cell-mediated cytotoxicity. Clin Cancer Res 2009; 15: 5108–5116.
Sauer M, Reiners KS, Hansen HP, Engert A, Gasser S, von Strandmann EP . Induction of the DNA damage response by IAP inhibition triggers natural immunity via upregulation of NKG2D ligands in Hodgkin lymphoma in vitro. Biol Chem 2013; 394: 1325–1331.
Heinemann A, Zhao F, Pechlivanis S, Eberle J, Steinle A, Diederichs S et al. Tumor suppressive micrornas MIR-34A/C control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res 2012; 72: 460–471.
Iannello A, Thompson TW, Ardolino M, Lowe SW, Raulet DH . P53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J Exp Med 2013; 210: 2057–2069.
Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A . Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res 2011; 71: 5998–6009.
Armeanu S, Bitzer M, Lauer UM, Venturelli S, Pathil A, Krusch M et al. Natural killer cell-mediated lysis of hepatoma cells via specific induction of NKG2D ligands by the histone deacetylase inhibitor sodium valproate. Cancer Res 2005; 65: 6321–6329.
Klein JM, Henke A, Sauer M, Bessler M, Reiners KS, Engert A et al. The histone deacetylase inhibitor LBH589 (panobinostat) modulates the crosstalk of lymphocytes with Hodgkin lymphoma cell lines. PLoS One 2013; 8: e79502.
Routes JM, Ryan S, Morris K, Takaki R, Cerwenka A, Lanier LL . Adenovirus serotype 5 E1A sensitizes tumor cells to NKG2D-dependent NK cell lysis and tumor rejection. J Exp Med 2005; 202: 1477–1482.
Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol 2010; 17: 471–482.
Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood 2006; 107: 4407–4416.
Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM . The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med 1988; 167: 353–371.
Kovalchuk AL, Qi CF, Torrey TA, Taddesse-Heath L, Feigenbaum L, Park SS et al. Burkitt lymphoma in the mouse. J Exp Med 2000; 192: 1183–1190.
Deng W, Gowen BG, Zhang L, Wang L, Lau S, Iannello A et al. Antitumor immunity. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science 2015; 348: 136–139.
Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity 2007; 27: 965–974.
Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M et al. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 2013; 121: 3658–3665.
Simhadri VR, Reiners KS, Hansen HP, Topolar D, Simhadri VL, Nohroudi K et al. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS One 2008; 3: e3377.
Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2015; 5: e1071008 eCollection 2016.
Lee JS . Activation of ATM-dependent DNA damage signal pathway by a histone deacetylase inhibitor, trichostatin A. Cancer Res Treat 2007; 39: 125–130.
Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A . Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK- cell lysis. Blood 2007; 110: 606–615.
Molinero LL, Fuertes MB, Girart MV, Fainboim L, Rabinovich GA, Costas MA et al. NF-kappa B regulates expression of the MHC class I-related chain A gene in activated T lymphocytes. J Immunol 2004; 173: 5583–5590.
Lin D, Lavender H, Soilleux EJ, O'Callaghan CA . NF-kappaB regulates MICA gene transcription in endothelial cell through a genetically inhibitable control site. J Biol Chem 2012; 287: 4299–4310.
Ashburner BP, Westerheide SD, Baldwin AS Jr . The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol 2001; 21: 7065–7077.
Dai Y, Rahmani M, Dent P, Grant S . Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol 2005; 25: 5429–5444.
Sato T, Kotake D, Hiratsuka M, Hirasawa N . Enhancement of inflammatory protein expression and nuclear factor kappaB (NF-kappaB) activity by trichostatin A (TSA) in OP9 preadipocytes. PLoS One 2013; 8: e59702.
Andresen L, Jensen H, Pedersen MT, Hansen KA, Skov S . Molecular regulation of MHC class I chain-related protein A expression after HDAC-inhibitor treatment of Jurkat T cells. J Immunol 2007; 179: 8235–8242.
Li H, Lakshmikanth T, Garofalo C, Enge M, Spinnler C, Anichini A et al. Pharmacological activation of p53 triggers anticancer innate immune response through induction of ULBP2. Cell Cycle 2011; 10: 3346–3358.
Soriani A, Iannitto ML, Ricci B, Fionda C, Malgarini G, Morrone S et al. Reactive oxygen species- and DNA damage response-dependent NK cell activating ligand upregulation occurs at transcriptional levels and requires the transcriptional factor E2F1. J Immunol 2014; 193: 950–960.
Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev 2000; 14: 272–277.
Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet 2000; 24: 300–303.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011; 471: 189–195.
Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 2012; 44: 1310–1315.
Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011; 471: 235–239.
Behera J, Jayprakash V, Sinha BN . Histone deacetylase inhibitors: a review on class-I specific inhibition. Mini Rev Med Chem 2015; 15: 731–750.
Schotterl S, Brennenstuhl H, Naumann U . Modulation of immune responses by histone deacetylase inhibitors. Crit Rev Oncog 2015; 20: 139–154.
Arrighetti N, Corno C, Gatti L . Drug combinations with HDAC inhibitors in antitumor therapy. Crit Rev Oncog 2015; 20: 83–117.
Acknowledgements
This study was supported by the Deutsche Forschungsgemeinschaft (KFO286, RP4; to EPvS) and the José Carreras-Leukämie Stiftung (DJCLS R 14/08; to EPvS). This study contains parts of the PhD thesis from MS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Sauer, M., Schuldner, M., Hoffmann, N. et al. CBP/p300 acetyltransferases regulate the expression of NKG2D ligands on tumor cells. Oncogene 36, 933–941 (2017). https://doi.org/10.1038/onc.2016.259
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.259
This article is cited by
-
Targeting natural killer cells: from basic biology to clinical application in hematologic malignancies
Experimental Hematology & Oncology (2024)
-
KLF4-mediated upregulation of the NKG2D ligand MICA in acute myeloid leukemia: a novel therapeutic target identified by enChIP
Cell Communication and Signaling (2023)
-
Induction of NKG2D ligand expression on tumor cells by CD8+ T-cell engagement-mediated activation of nuclear factor-kappa B and p300/CBP-associated factor
Oncogene (2019)