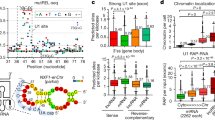
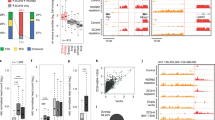
Abstract
Genome-wide chromatin studies identified the tumor suppressor p53 as both a promoter and an enhancer-binding transcription factor. As an enhancer factor, p53 can induce local production of enhancer RNAs, as well as transcriptional activation of distal neighboring genes. Beyond the regulation of protein-coding genes, p53 has the capacity to regulate long intergenic noncoding RNA molecules (lincRNAs); however, their importance to the p53 tumor suppressive function remains poorly characterized. Here, we identified and characterized a novel p53-bound intronic enhancer that controls the expression of its host, the lincRNA00475 (linc-475). We demonstrate the requirement of linc-475 for the proper induction of a p53-dependent cell cycle inhibitory response. We further confirm the functional importance of linc-475 in the maintenance of CDKN1A/p21 levels, a cell cycle inhibitor and a major p53 target gene, following p53 activation. Interestingly, loss of linc-475 reduced the binding of both p53 and RNA polymerase II (RNAPII) to the promoter of p21, attenuating its transcription rate following p53 activation. Altogether, our data suggest a direct role of p53-bound enhancer domains in the activation of lincRNAs required for an efficient p53 transcriptional response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lane DP . Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16.
Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 2011; 145: 571–583.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142: 409–419.
Léveillé N, Melo CA, Rooijers K, Lagares AD, Melo SA, KorkMaz G et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun 2015; 6: 6520.
De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010; 8: e1000384.
Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465: 182–187.
Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 2013; 49: 524–535.
Rinn JL, Chang HY . Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–166.
Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 2012; 19: 1992–2002.
Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011; 473: 43–49.
Splinter E, de Laat W . The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J 2011; 30: 4345–4355.
Dekker J, Rippe K, Dekker M, Kleckner N . Capturing chromosome conformation. Science 2002; 295: 1306–1311.
Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 2011; 25: 1371–1383.
Rippe K . Making contacts on a nucleic acid polymer. Trends Biochem Sci 2001; 26: 733–740.
Phillips JE, Corces VG . CTCF: master weaver of the genome. Cell 2009; 137: 1194–1211.
Zhou X, Lowdon RF, Li D, Lawson HA, Madden PA, Costello JF et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nat Methods 2013; 10: 375–376.
Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK . Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 2014; 56: 29–42.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485: 376–380.
Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 2014; 111: 996–1001.
Agarwal ML, Agarwal A, Taylor WR, Stark GR . p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA 1995; 92: 8493–8497.
Cho SW, Kim S, Kim JM, Kim JS . Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31: 230–232.
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013; 154: 1380–1389.
Sanjana NE, Shalem O, Zhang F . Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11: 783–784.
Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW . Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88: 593–602.
Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell 2013; 12: 890–900.
Kumar PP, Emechebe U, Smith R, Franklin S, Moore B, Yandell M et al. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. eLife 2014, e-pub ahead of print 29 May 2014 doi:10.7554/eLife.02805.
Lazorthes S, Vallot C, Briois S, Aguirrebengoa M, Thuret JY, Laurent GS et al. A vlincRNA participates in senescence maintenance by relieving H2AZ-mediated repression at the INK4 locus. Nat Commun 2015; 6: 5971.
Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006; 124: 1169–1181.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014; 26: 344–357.
Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 2013; 14: R104.
Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 2014; 3: e02200.
Grossi E, Sanchez Y, Huarte M . Expanding the p53 regulatory network: LncRNAs take up the challenge. Biochim Biophys Acta 2015, ; e-pub ahead of print 18 July 2015 doi:10.1016/j.bbagrm.2015.07.011.
Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013; 498: 516–520.
Shlyueva D, Stampfel G, Stark A . Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 2014; 15: 272–286.
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY . Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011; 44: 667–678.
Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 2014; 5: 5812.
Leeuw JD, Hornik K, Mair P . Isotone optimization in R: pool-adjacent-violators algorithm (PAVA) and active set methods. J Stat Softw 2009; 32: 1–24.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Melo, C., Léveillé, N., Rooijers, K. et al. A p53-bound enhancer region controls a long intergenic noncoding RNA required for p53 stress response. Oncogene 35, 4399–4406 (2016). https://doi.org/10.1038/onc.2015.502
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.502
This article is cited by
-
Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370-3p/FGF14 axis
Cell Death Discovery (2020)
-
Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer
Journal of Hematology & Oncology (2017)