Abstract
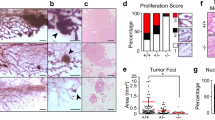
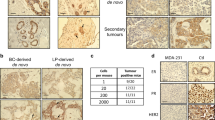
Separase, a protease encoded by the ESPL1 gene, cleaves the chromosomal cohesin during mitosis. Separase protein and transcripts are overexpressed in a wide range of human cancers. To investigate the physiological consequence of Separase overexpression in animals, we have generated a transgenic MMTV-Espl1 mouse model that overexpresses Separase protein in the mammary glands. MMTV-Espl1 mice in a C57BL/6 genetic background develop aggressive, highly aneuploid and estrogen receptor alpha-positive (ERα+) mammary adenocarcinomas with an 80% penetrance. The mammary tumors caused by overexpression of Separase, alone or combined with p53 heterozygosity, in mammary epithelium mimic several aspects of the most aggressive forms of human breast cancer, including high levels of genetic instability, cell cycle defects, poor differentiation, distant metastasis and metaplasia. Histopathologically, MMTV-Espl1 tumors are highly heterogeneous showing features of both luminal as well as basal subtypes of breast cancers, with aggressive disease phenotype. In addition to aneuploidy, Separase overexpression results in chromosomal instability (CIN) including premature chromatid separation (PCS), lagging chromosomes, anaphase bridges, micronuclei, centrosome amplification, multinucleated cells, gradual accumulation of DNA damage and progressive loss of tumor suppressors p53 and cadherin gene loci. These results suggest that Separase-overexpressing mammary cells are not only susceptible to chromosomal missegregation-induced aneuploidy but also other genetic instabilities including DNA damage and loss of key tumor suppressor gene loci, which in combination can initiate tumorigenesis and disease progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Nasmyth K, Haering CH . Cohesin: its roles and mechanisms. Annu Rev Genet 2009; 43: 525–558.
Meyer R, Fofanov V, Panigrahi AK, Merchant F, Zhang N, Pati D . Overexpression and mislocalizaion of the chromosomal segregation protein Separase in multiple human cancers. Clin Cancer Res 2009; 15: 2703–2710.
Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA 2008; 105: 13033–13038.
Duesberg P, Rausch C, Rasnick D, Hehlmann R . Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA 1998; 95: 13692–13697.
Holland AJ, Cleveland DW . Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 2009; 10: 478–487.
Holland AJ, Cleveland DW . Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep 2012; 13: 501–514.
McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C . Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 2012; 13: 528–538.
Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA 2007; 104: 8995–9000.
Heilig CE, Loffler H, Mahlknecht U, Janssen JW, Ho AD, Jauch A et al. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med 2010; 14: 895–902.
Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997; 57: 1597–1604.
McClelland SE, Burrell RA, Swanton C . Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle 2009; 8: 3262–3266.
Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci USA 2009; 106: 8671–8676.
Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007; 317: 916–924.
Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 2008; 322: 703–709.
Sheltzer JM, Amon A . The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet 2011; 27: 446–453.
Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010; 468: 321–325.
Pariente N . A balancing act: focus on aneuploidy. EMBO Rep 2012; 13: 472.
Berman J . Evolutionary genomics: When abnormality is beneficial. Nature 2010; 468: 183–184.
Ornitz DM, Moreadith RW, Leder P . Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci USA 1991; 88: 698–702.
Blackburn AC, McLary SC, Naeem R, Luszcz J, Stockton DW, Donehower LA et al. Loss of heterozygosity occurs via mitotic recombination in Trp53+/− mice and associates with mammary tumor susceptibility of the BALB/c strain. Cancer Res 2004; 64: 5140–5147.
Yamamoto KR . Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet 1985; 19: 209–252.
Sonnenberg A, van BP, Hilgers J, Schuuring E, Nusse R . Oncogene expression during progression of mouse mammary tumor cells; activity of a proviral enhancer and the resulting expression of int-2 is influenced by the state of differentiation. EMBO J 1987; 6: 121–125.
Arnold A, Papanikolaou A . Cyclin D1 in breast cancer pathogenesis. J Clin Oncol 2005; 23: 4215–4224.
Lacroix M, Leclercq G . About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol 2004; 219: 1–7.
Lacroix M, Toillon RA, Leclercq G . Stable 'portrait' of breast tumors during progression: data from biology, pathology and genetics. Endocr Relat Cancer 2004; 11: 497–522.
Cariou S, Donovan JC, Flanagan WM, Milic A, Bhattacharya N, Slingerland JM . Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc Natl Acad Sci USA 2000; 97: 9042–9046.
Musgrove EA, Sutherland RL . Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009; 9: 631–643.
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 2008; 283: 29897–29903.
Han S, Park K, Kim HY, Lee MS, Kim HJ, Kim YD . Reduced expression of p27Kip1 protein is associated with poor clinical outcome of breast cancer patients treated with systemic chemotherapy and is linked to cell proliferation and differentiation. Breast Cancer Res Treat 1999; 55: 161–167.
Cardiff RD . The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 2010; 15: 225–233.
Fantozzi A, Christofori G . Mouse models of breast cancer metastasis. Breast Cancer Res 2006; 8: 212.
Cancer Genome Atlas Network, Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 2009; 106: 13820–13825.
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8: R76.
Janssen A, van der BM, Szuhai K, Kops GJ, Medema RH . Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011; 333: 1895–1898.
Lu X, Yang C, Yin C, Van DT, Simin K . Apoptosis is the essential target of selective pressure against p53, whereas loss of additional p53 functions facilitates carcinoma progression. Mol Cancer Res 2011; 9: 430–439.
Pati D, Haddad BR, Haegele A, Thompson H, Kittrell FS, Shepard A et al. Hormone-induced chromosomal instability in p53-null mammary epithelium. Cancer Res 2004; 64: 5608–5616.
Ganmore I, Smooha G, Izraeli S . Constitutional aneuploidy and cancer predisposition. Hum Mol Genet 2009; 18: R84–R93.
Panigrahi AK, Pati D . Road to the crossroads of life and death: Linking sister chromatid cohesion and separation to aneuploidy, apoptosis and cancer. Crit Rev Oncol Hematol 2009; 72: 181–193.
Solomon DA, Kim T, az-Martinez LA, Fair J, Elkahloun AG, Harris BT et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 2011; 333: 1039–1043.
Shepard JL, Amatruda JF, Finkelstein D, Ziai J, Finley KR, Stern HM et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev 2007; 21: 55–59.
Stegmeier F, Visintin R, Amon A . Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 2002; 108: 207–220.
Mukherjee M, Ge G, Zhang N, Huang E, Nakamura LV, Minor M et al. Separase loss of function cooperates with the loss of p53 in the initiation and progression of T- and B-cell lymphoma, leukemia and aneuploidy in mice. PLoS ONE 2011; 6: e22167.
Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM . Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013; 18: 123–133.
Radaelli E, Damonte P, Cardiff RD . Epithelial-mesenchymal transition in mouse mammary tumorigenesis. Future Oncol 2009; 5: 1113–1127.
Stefansson OA, Jonasson JG, Johannsson OT, Olafsdottir K, Steinarsdottir M, Valgeirsdottir S et al. Genomic profiling of breast tumours in relation to BRCA abnormalities and phenotypes. Breast Cancer Res 2009; 11: R47.
Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992; 258: 818–821.
Medina D, Kittrell FS . ‘Establishment of Mouse Mammary Cell Lines,’in Methods in mammary Gland Biology and Breast Cancer Research Ip MM and Asch BB (eds) Kluwer Academic Press, New York, NY, USA, 2000, pp 137–145.
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009; 27: 1160–1167.
Acknowledgements
This study was supported by the grants from the National Cancer Institute (1RO1 CA109330 and 1RO1 CA109478) to D. Pati.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Mukherjee, M., Ge, G., Zhang, N. et al. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ERα)-positive mammary adenocarcinomas. Oncogene 33, 5511–5522 (2014). https://doi.org/10.1038/onc.2013.493
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.493
Keywords
This article is cited by
-
Genome-wide CRISPR screen identifies ESPL1 limits the response of gastric cancer cells to apatinib
Cancer Cell International (2024)
-
Let-7c-5p Restrains Cell Growth and Induces Apoptosis of Lung Adenocarcinoma Cells via Targeting ESPL1
Molecular Biotechnology (2022)
-
Aneuploidy as a promoter and suppressor of malignant growth
Nature Reviews Cancer (2021)
-
Centrosome reduction in newly-generated tetraploid cancer cells obtained by separase depletion
Scientific Reports (2020)
-
Cytoplasmic ERα and NFκB Promote Cell Survival in Mouse Mammary Cancer Cell Lines
Hormones and Cancer (2020)