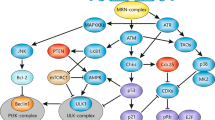
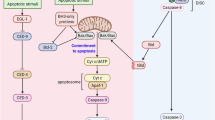
Abstract
The consequences of DNA damage depend on the cell type and the severity of the damage. Mild DNA damage can be repaired with or without cell-cycle arrest. More severe and irreparable DNA injury leads to the appearance of cells that carry mutations or causes a shift towards induction of the senescence or cell death programs. Although for many years it was argued that DNA damage kills cells via apoptosis or necrosis, technical and methodological progress during the last few years has helped to reveal that this injury might also activate death by autophagy or mitotic catastrophe, which may then be followed by apoptosis or necrosis. The molecular basis underlying the decision-making process is currently the subject of intense investigation. Here, we review current knowledge about the response to DNA damage and subsequent signaling, with particular attention to cell death induction and the molecular switches between different cell death modalities following damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ward I, Chen J . Early events in the DNA damage response. Curr Top Dev Biol 2004; 63: 1–35.
Wang B, Matsuoka S, Carpenter PB, Elledge SJ . 53BP1, a mediator of the DNA damage checkpoint. Science 2002; 298: 1435–1438.
Stucki M, Jackson SP . MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst) 2004; 3: 953–957.
Paull TT, Lee JH . The Mre11/Rad50/Nbs1 complex and its role as a DNA double-strand break sensor for ATM. Cell Cycle 2005; 4: 737–740.
Pallis AG, Karamouzis MV . DNA repair pathways and their implication in cancer treatment. Cancer Metastasis Rev 2010; 29: 677–685.
Freeman AK, Monteiro AN . Phosphatases in the cellular response to DNA damage. Cell Commun Signaling 2010; 8: 27–38.
Lane DP . Cancer. p53, guardian of the genome. Nature 1992; 358: 15–16.
Bartek J, Lukas J . Pathways governing G1/S transition and their response to DNA damage. FEBS Lett 2001; 490: 117–122.
Brown JP, Wei W, Sedivy JM . Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science 1997; 277: 831–834.
Campisi J, D’Adda di Fagagna F . Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 2007; 8: 729–740.
Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC . Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA 1996; 93: 13742–13747.
Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 2003; 22: 4212–4222.
Saretzki G . Cellular senescence in the development and treatment of cancer. Curr Pharm Des 2010; 16: 79–100.
Webley K, Bond JA, Jones CJ, Blaydes JP, Craig A, Hupp T . Posttranslational modifications of p53 in replicative senescence overlapping but distinct from those induced by DNA damage. Mol Cell Biol 2000; 20: 2803–2808.
Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM . A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 2002; 109: 335–346.
Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849–862.
Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge WH et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 2002; 4: 1–10.
D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S et al. Homeodomaininteracting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4: 11–19.
Gresko E, Roscic A, Ritterhoff S, Vichalkovski A, Del Sal G, Schmitz ML . Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J 2006; 25: 1883–1894.
Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F et al. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 2007; 25: 739–750.
Hock AK, Vousden KH . Tumor suppression by p53: fall of the triumvirate? Cell 2012; 149: 1183–1185.
Kim H, Lee JM, Lee G, Bhin J, Oh SK, Kim K et al. DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Mol Cell 2011; 44: 797–810.
He L, He X, Lim LP, Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007; 26: 731–743.
Jin Z, El-Deiry WS . Overview of cell death signaling pathways. Cancer Biol Ther 2005; 4: 139–163.
Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002; 416: 560–564.
Sayan AE, Sayan BS, Gogvadze V, Dinsdale D, Nyman U, Hansen TM et al. P73 and caspase cleaved p73 fragments localize to mitochondria and augment TRAIL-induced apoptosis. Oncogene 2008; 27: 4363–4372.
Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science 2000; 289: 1159–1164.
De Le’se’leuc L, Denis F . Inhibition of apoptosis by Nur77 through NF-kB activity modulation. Cell Death Differ 2006; 13: 293–300.
Tinel A, Tschopp J . The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 2004; 304: 843–846.
Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S et al. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ 2012; 8: 1288–1298.
Vakifahmetoglu H, Olsson M, Tamm C, Heidari N, Orrenius S, Zhivotovsky B . DNA damage induces two distinct modes of cell death in ovarian carcinomas. Cell Death Differ 2008; 15: 555–566.
Castedo M, Perfettini JL, Roumier T, Yakushijin K, Horne D, Medema R . The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene 2004; 23: 4353–4361.
Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB . p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 2007; 11: 175–189.
Waldman T, Lengauer C, Kinzler KW, Vogelstein B . Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 1996; 381: 713–716.
Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G . Cell death by mitotic catastrophe: a molecular definition. Oncogene 2004; 236: 2825–2837.
Vakifahmetoglu H, Olsson M, Zhivotovsky B . Death through a tragedy: mitotic catastrophe. Cell Death Differ 2008; 15: 1153–1162.
Vitale I, Galluzzi L, Castedo M, Kroemer G . Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011; 6: 385–392.
Zeng X, Kinsella TJ . BNIP3 is essential for mediating 6-thioguanineand 5-fluorouracil-induced autophagy following DNA mismatch repair processing. Cell Res 2010; 20: 665–675.
Feng Z, Zhang H, Levine AJ, Jin S . The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 2005; 102: 8204–8209.
Levine B, Abrams J . p53: the Janus of autophagy? Nat Cell Biol 2008; 10: 637–639.
Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10: 676–687.
Alexander A, Kim J, Walker CL . ATM engages the TSC2/mTORC1 signaling node to regulate autophagy. Autophagy 2010; 6: 672–673.
Muñoz-Gámez JA, Rodríguez-Vargas JM, Quiles-Pérez R, Aguilar-Quesada R, Martín-Oliva D, Murcia G et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy 2009; 5: 61–74.
Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471: 74–79.
Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB et al. Role of Bcl-2 family proteins in a nonapoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 2004; 6: 1221–1228.
Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH . PUMA and Bax-induced autophagy contributes to apoptosis. Cell Death Differ 2009; 16: 1135–1145.
Lorin S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M . Evidence for the interplay between JNK and p53-DRAM signaling pathways in the regulation of autophagy. Autophagy 2010; 6: 153–154.
Berger NA . Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res 1985; 101: 4–15.
Liaudet L, Soriano FG, Szabó E, Virág L, Mabley JG, Salzman AL et al. Protection against hemorrhagic shock in mice genetically deficient in poly(ADP-ribose)polymerase. Proc Natl Acad Sci USA 2000; 97: 10203–10208.
Virág L, Scott GS, Cuzzocrea S, Marmer D, Salzman AL, Szabó C . Peroxynitrite-induced thymocyte apoptosis: the role of caspases and poly (ADP-ribose) synthetase (PARS) activation. Immunology 1998; 94: 345–355.
Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG . Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res 1993; 53: 3976–3985.
Menissier-de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA 1997; 94: 7303–7307.
Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem 1998; 23: 14461–14467.
Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K . Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J 2005; 24: 1375–1386.
Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG . Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem 2005; 280: 6447–6454.
Norberg E, Karlsson M, Korenovska O, Szydlowski S, Silberberg G, Uhlén P et al. Critical role for hyperpolarization-activated cyclic nucleotide-gated channel 2 in the AIF-mediated apoptosis. EMBO J 2010; 22: 3869–3878.
Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J et al. Sequential activation of poly(ADPRibose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol 2007; 27: 4844–4862.
Artus C, Boujrad H, Bouharrour A, Brunelle MN, Hoos S, Yuste VJ et al. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J 2010; 9: 1585–1599.
Bai S, Goodrich DW . Different DNA lesions trigger distinct cell death responses in HCT116 colon carcinoma cells. Mol Cancer Ther 2004; 3: 613–620.
Vakifahmetoglu H, Olsson M, Tamm C, Heidari N, Orrenius S, Zhivotovsky B . DNA damage induces two distinct modes of cell death in ovarian carcinomas. Cell Death Differ 2008; 15: 555–566.
Borges HL, Linden R, Wang JYJ . DNA damage-induced cell death: lessons from the central nervous system. Cell Res 2008; 18: 17–26.
Wang JY, Cho SK . Coordination of repair, checkpoint, and cell death responses to DNA damage. Adv Protein Chem 2004; 69: 101–135.
Nelyudova A, Aksenov ND, Pospelov V, Pospelova T . By blocking apoptosis, Bcl-2 in p38-dependent manner promotes cell cycle arrest and accelerated senescence after DNA damage and serum withdrawal. Cell Cycle 2007; 6: 2171–2177.
Rebbaa A, Zheng X, Chou PM, Mirkin BL . Caspase inhibition switches doxorubicin-induced apoptosis to senescence. Oncogene 2003; 18: 2805–2811.
Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y . Autophagy mediates the process of cellular senescence characterizing bile duct damages in primary biliary cirrhosis. Lab Invest 2010; 90: 835–843.
Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF et al. Autophagy mediates the mitotic senescence transition. Genes Dev 2009; 23: 798–803.
Abedin MJ, Wang D, McDonnell MA, Lehmann U, Keleka A . Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 2007; 14: 500–510.
Kaminskyy V, Piskunova T, Zborovskaya I, Tchevkina E, Zhivotovsky B . Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy 2012; 8: 1032–1044.
Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012; 8: 810–820.
Samara C, Syntichaki P, Tavernarakis N . Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ 2008; 15: 105–112.
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 1: 51–64.
Morandell S, Yaffe MB . Exploiting synthetic lethal interactions between DNA damage signaling, checkpoint control, and p53 for targeted cancer therapy. Prog Mol Biol 2012; 110: 289–314.
Acknowledgements
The work in the author’s laboratory was supported by grants from the Swedish Research Council, the Swedish and the Stockholm Cancer Societies, the Swedish Childhood Cancer Foundation, and the FP7 (Apo-Sys) program. OS was supported by a fellowship from the Swedish Institute and Karolinska Institutet. We apologize to those authors whose primary references could not be cited due to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Surova, O., Zhivotovsky, B. Various modes of cell death induced by DNA damage. Oncogene 32, 3789–3797 (2013). https://doi.org/10.1038/onc.2012.556
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.556
Keywords
This article is cited by
-
Nasopharyngeal carcinoma: current views on the tumor microenvironment's impact on drug resistance and clinical outcomes
Molecular Cancer (2024)
-
TRIM24 is critical for the cellular response to DNA double-strand breaks through regulating the recruitment of MRN complex
Oncogene (2023)
-
Emerging and established therapies for chemotherapy-induced ototoxicity
Journal of Cancer Survivorship (2023)
-
Sirtuin5 protects colorectal cancer from DNA damage by keeping nucleotide availability
Nature Communications (2022)
-
Radiobiological effects of wound fluid on breast cancer cell lines and human-derived tumor spheroids in 2D and microfluidic culture
Scientific Reports (2022)