Abstract
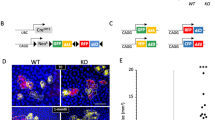
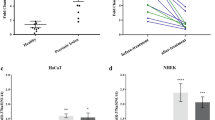
Epidermal differentiation and stratification, crucial for barrier formation, are regulated by a complex interplay of transcription factors, including the evolutionarily conserved Grainyhead-like 3 (Grhl3/Get1); Grhl3-deleted mice exhibit impaired epidermal differentiation and decreased expression of multiple differentiation genes. To test whether Grhl3 regulates epidermal genes indirectly by controlling the expression of specific microRNAs (miRs), we performed miR profiling and identified 11 miRs that are differentially regulated in Grhl3−/− skin, one of which is miR-21, previously shown to be upregulated in diseased skin, including in psoriasis and squamous cell skin cancer. We found that miR-21 is normally expressed in the post-mitotic suprabasal layers of the epidermis, overlapping with Grhl3. The miR-21 promoter is bound and repressed by Grhl3 indicating that these two factors are involved in a regulatory loop maintaining homeostasis in the epidermis. Although miR-21 overexpression in normal keratinocytes had mild effects on the expression of several known miR-21 targets, an enhanced downregulation of the miR-21 tumor-related targets, including MSH2, was observed in Ras-transformed keratinocytes. The increased sensitivity of transformed keratinocytes to miR-21’s effects occurs in part through downregulation of the RNA-binding protein DND1 during the transformation process. Additionally, we observed increased tumorigenesis in mice subcutaneously injected with transformed keratinocytes lacking Grhl3. These findings indicate that decreased Grhl3 expression contributes to tumor progression and upregulation of the oncomir miR-21 in squamous cell carcinoma of the skin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Koster MI, Roop DR . Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol 2007; 23: 93–113.
Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 2006; 16: 1041–1049.
Yi R, Poy MN, Stoffel M, Fuchs E . A skin microRNA promotes differentiation by repressing 'stemness'. Nature 2008; 452: 225–229.
Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA 2009; 106: 498–502.
Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G et al. miR-203 represses 'stemness' by repressing DeltaNp63. Cell Death Differ 2008; 15: 1187–1195.
Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C . Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 2010; 130: 1249–1257.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10: 593–601.
Korpal M, Lee ES, Hu G, Kang Y . The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283: 14910–14914.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH . miR-200b mediates post-transcriptional repression of ZFHX1B. RNA 2007; 13: 1172–1178.
Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 2005; 308: 411–413.
Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol 2006; 299: 122–136.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H et al. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol 2010; 130: 124–134.
Hildebrand J, Rutze M, Walz N, Gallinat S, Wenck H, Deppert W et al. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol 2011; 131: 20–29.
Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 2006; 38: 356–362.
Ahmed MI, Mardaryev AN, Lewis CJ, Sharov AA, Botchkareva NV . MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J Cell Sci 2011; 124: 3399–3404.
Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 2011; 20: 4025–4040.
Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2007; 2: e610.
Zibert JR, Lovendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L . MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci 2010; 58: 177–185.
Dziunycz P, Iotzova-Weiss G, Eloranta JJ, Lauchli S, Hafner J, French LE et al. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J Invest Dermatol 2010; 130: 2686–2689.
Ting SB, Caddy J, Wilanowski T, Auden A, Cunningham JM, Elias PM et al. The epidermis of grhl3-null mice displays altered lipid processing and cellular hyperproliferation. Organogenesis 2005; 2: 33–35.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Sandelin A, Wasserman WW, Lenhard B . ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res 2004; 32: W249–W252.
Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 2008; 378: 492–504.
Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008; 452: 896–899.
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 2005; 438: 685–689.
Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010; 18: 282–293.
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500.
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: P3.
Sheibani N, Rhim JS, Allen-Hoffmann BL . Malignant human papillomavirus type 16-transformed human keratinocytes exhibit altered expression of extracellular matrix glycoproteins. Cancer Res 1991; 51: 5967–5975.
Jiang R, Cabras G, Sheng W, Zeng Y, Ooka T . Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia 2009; 11: 964–973.
Meira LB, Cheo DL, Reis AM, Claij N, Burns DK, te Riele H et al. Mice defective in the mismatch repair gene Msh2 show increased predisposition to UVB radiation-induced skin cancer. DNA Repair (Amst) 2002; 1: 929–934.
Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H et al. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res 1996; 56: 3842–3849.
Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007; 131: 1273–1286.
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W . Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006; 125: 1111–1124.
Zhu R, Iacovino M, Mahen E, Kyba M, Matin A . Transcripts that associate with the RNA binding protein, DEAD-END (DND1), in embryonic stem (ES) cells. BMC Mol Biol 2011; 12: 37.
Selcuklu SD, Donoghue MT, Spillane C . miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 2009; 37: 918–925.
Yu Y, Wang Y, Ren X, Tsuyada A, Li A, Liu LJ et al. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor beta contributes to chemoresistance in breast cancer cells. Mol Cancer Res 2010; 8: 1633–1642.
Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q et al. Targeting of the tumor suppressor GRHL3 by a miR-21-Dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011; 20: 635–648.
Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci USA 2011; 108: 10144–10149.
Wang Y, Lee CG . MicroRNA and cancer —focus on apoptosis. J Cell Mol Med 2009; 13: 12–23.
Ribas J, Lupold SE . The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle 2010; 9: 923–929.
Cottonham CL, Kaneko S, Xu L . miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem 2010; 285: 35293–35302.
Yang X, Wang J, Guo SL, Fan KJ, Li J, Wang YL et al. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int J Biol Sci 2011; 7: 685–690.
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K . STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010; 39: 493–506.
Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J 1999; 18: 4657–4668.
Kira M, Sano S, Takagi S, Yoshikawa K, Takeda J, Itami S . STAT3 deficiency in keratinocytes leads to compromised cell migration through hyperphosphorylation of p130(cas). J Biol Chem 2002; 277: 12931–12936.
Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P et al. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet 2009; 5: e1000573.
Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ . E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 2008; 18: 1763–1777.
Roop DR, Lowy DR, Tambourin PE, Strickland J, Harper JR, Balaschak M et al. An activated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature 1986; 323: 822–824.
Acknowledgements
We thank Stuart Yuspa and Andrew Ryscavage for the gift of mouse papilloma and carcinoma sample and the Ras virus. We also thank Reuven Agami for HA-DND1 expression plasmid. This work was supported by grants from the NIH (AR44882) and the Irving Weinstein Foundation (to BA); the California Tobacco Related Disease Research Program (17DT-0192 to AB); the NIH training program in Systems Biology of Development (T32-HD60555 to WG); the National Library of Medicine training grant (LM07443 to EG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Bhandari, A., Gordon, W., Dizon, D. et al. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene 32, 1497–1507 (2013). https://doi.org/10.1038/onc.2012.168
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.168
Keywords
This article is cited by
-
Gene expression in tonsils in swine following infection with porcine reproductive and respiratory syndrome virus
BMC Veterinary Research (2021)
-
The RNA–RNA base pairing potential of human Dicer and Ago2 proteins
Cellular and Molecular Life Sciences (2020)
-
PCBP1 depletion promotes tumorigenesis through attenuation of p27Kip1 mRNA stability and translation
Journal of Experimental & Clinical Cancer Research (2018)
-
Stage-dependent therapeutic efficacy in PI3K/mTOR-driven squamous cell carcinoma of the skin
Cell Death & Differentiation (2018)
-
Roles of Grainyhead-like transcription factors in cancer
Oncogene (2017)