Abstract
The stromal microenvironment has key roles in prostate development and cancer, and cancer-associated fibroblasts (CAFs) stimulate tumourigenesis via several mechanisms including the expression of pro-tumourigenic factors. Mesenchyme (embryonic stroma) controls prostate organogenesis, and in some circumstances can re-differentiate prostate tumours. We have applied next-generation Tag profiling to fetal human prostate, normal human prostate fibroblasts (NPFs) and CAFs to identify molecules expressed in prostatic stroma. Comparison of gene expression profiles of a patient-matched pair of NPFs vs CAFs identified 671 transcripts that were enriched in CAFs and 356 transcripts whose levels were decreased, relative to NPFs. Gene ontology analysis revealed that CAF-enriched transcripts were associated with prostate morphogenesis and CAF-depleted transcripts were associated with cell cycle. We selected mRNAs to follow-up by comparison of our data sets with published prostate cancer fibroblast microarray profiles as well as by focusing on transcripts encoding secreted and peripheral membrane proteins, as well as mesenchymal transcripts identified in a previous study from our group. We confirmed differential transcript expression between CAFs and NPFs using QrtPCR, and defined protein localization using immunohistochemistry in fetal prostate, adult prostate and prostate cancer. We demonstrated that ASPN, CAV1, CFH, CTSK, DCN, FBLN1, FHL1, FN, NKTR, OGN, PARVA, S100A6, SPARC, STC1 and ZEB1 proteins showed specific and varied expression patterns in fetal human prostate and in prostate cancer. Colocalization studies suggested that some stromally expressed molecules were also expressed in subsets of tumour epithelia, indicating that they may be novel markers of EMT. Additionally, two molecules (ASPN and STC1) marked overlapping and distinct subregions of stroma associated with tumour epithelia and may represent new CAF markers.
Similar content being viewed by others
Introduction
Prostate cancer is the most prevalent cancer in UK men, with around 36 000 new cases and ∼10 000 deaths per year. The role of the tumour microenvironment in prostate cancer initiation and progression is important, but is poorly understood at the mechanistic or molecular level. Tumour stroma (cancer-associated fibroblasts, CAFs) stimulates prostate carcinogenesis (Olumi et al., 1999; Hayward et al., 2001) and stromal histology is an independent predictor of prostate cancer prognosis (Ayala et al., 2003; Yanagisawa et al., 2007). There is considerable interest in identifying pathways in tumour stroma that act as paracrine regulators of tumour epithelia, as these may become new therapeutics or diagnostics and would be a novel intervention in tumour growth. The study of stromal involvement in breast cancer has identified several functions in tumour progression and metastasis. Serial analysis of gene expression (SAGE) profiling of breast cancer stroma identified SDF1/CXCL12 (Allinen et al., 2004), which was subsequently shown to stimulate epithelial proliferation as well as recruitment of endothelial precursors from bone marrow (Orimo et al., 2005). Stromal gene expression pattern predicts disease progression (Finak et al., 2008; Ma et al., 2009), while signalling from mesenchymal stem cells via CCL5 stimulates metastasis (Karnoub et al., 2007). In the prostate, stromal signalling may initiate some tumours (Bhowmick et al., 2004; Placencio et al., 2008), stimulate tumour cell proliferation (Olumi et al., 1999; Hayward et al., 2001), enhance vasculogenesis (Tuxhorn et al., 2002) and control metastasis (Thalmann et al., 2010). TGFβ, CXCL12 and CXCL14 have been identified as pro-tumourigenic mediators made by prostate tumour stroma (Ao et al., 2007; Augsten et al., 2009).
The developing prostate shares some characteristics with prostate tumours, including rapid growth driven by paracrine interactions from the mesenchyme/stroma (Hayward et al., 1996). Embryonic mesenchyme has been shown to inhibit tumour growth in the Dunning tumour model system (Hayashi and Cunha, 1991), which illustrates the potency of mesenchymal paracrine pathways. Additionally, profiling of developmental mesenchyme has identified pathways that are dysregulated in CAFs (Vanpoucke et al., 2007), and other studies have demonstrated similarities between developmental gene expression and prostate tumours (Joesting et al., 2005; Schaeffer et al., 2008; Pritchard et al., 2009). CAFs are defined by their pro-tumourigenic signalling to epithelia, and assayed by tumour reconstitution studies in vivo and there appears to be significant cellular heterogeneity within CAFs (Sugimoto et al., 2006). Recent studies have demonstrated that the heterogeneity of tumour stroma and CAFs is a key feature of their pro-tumourigenic activity (Franco et al., 2011; Kiskowski et al., 2011). Gene profiling studies have examined the transcriptional profile of cancer stroma and stromal subsets (Zhao et al., 2007; Dakhova et al., 2009; Pascal et al., 2009) to identify CAF upregulated and downregulated molecules. The cellular heterogeneity of CAFs is a potential problem in gene profiling studies, since there may be a sub-population of cells that express key paracrine factors that are present as a minority among a complex mix of other cells. In regard to developmental studies, restricted subsets of prostatic mesenchyme are known to contain organ-inductive activity as well as localized expression of key regulatory molecules (Timms et al., 1995; Thomson and Cunha, 1999). By comparing transcriptional profiles of embryonic mesenchyme and CAFs, it may be possible to identify molecules expressed in cell subsets that would be poorly represented due to their dilution by transcripts from other cell types, as well molecules that are co-expressed due to the similarity of cellular events in development and disease. Furthermore, few molecules have been identified whose expression is restricted to the stromal compartment, and such molecules may be important as CAF markers or regulatory factors. In addition, the identification of stromal-specific molecules is a prerequisite for the identification of epithelial-mesenchymal transition (EMT) markers.
We have examined the transcripts expressed in CAF, normal human prostate fibroblast (NPF) and fetal prostate using Tag profiling, and compared our data set with published studies of prostate cancer stroma. This led to the identification of several putative stromally expressed transcripts, and expression was examined by QrtPCR in several matched CAF/NPF pairs and fetal prostate. The distribution of 15 candidate proteins was examined by immunohistochemistry in fetal human prostate, benign prostate and prostate cancer samples to verify stromal expression and to look for expression in stromal subsets or epithelia. Four molecules were further studied using confocal colocalization, to determine their co-expression in epithelia or stromal subsets. We identified several molecules expressed in developing human prostate mesenchyme and CAFs, whose expression was restricted to the stromal compartment, and our data are consistent with the paradigm that cancer and developmental systems share several pathways.
Results
Tag profiling of fetal human prostate, primary adult normal and cancer-associated prostate fibroblast cells
To identify transcripts expressed in normal and cancer prostate stroma and developing prostate, we performed Tag profiling of primary human normal and cancer-associated prostatic fibroblast cells (from the same patient) and human fetal (14–15 weeks) prostate. The gene expression profiles that we obtained by tag profiling yielded 2.4–3.6 million signature sequences (Tags) per data set for the NPF, CAF and fetal (embryonic, EMB) samples (available at GEO GSE25018). Each tag library was filtered to include only well-annotated tags (assigned class 1, 2 or 3 according to their relationship to the polyA tail). Next, the tag libraries were filtered to include tags with transcripts per million (TPM) ⩾3, which represents ∼1 transcript per cell, and eliminates extremely low frequency Tags. An overview of the composition of the CAF, NPF and EMB tag libraries is presented in Table 1.
To identify shared and unique tag expression in the NPF, CAF and EMB data sets, the libraries were compared by the tag/signature sequences using Microsoft Access software. Distinct and overlapping tag expression in the NPF, CAF and EMB libraries were determined, and are presented in a Venn diagram (Figure 1a). The numbers in the Venn diagram refer to tag number and not unique transcript expression (unigene ID) since there may be more than one tag type per gene due to splice variants. The number of unique tags in the EMB library was greater than the CAF and NPF libraries. The EMB tag library is derived from a complex tissue made up of several cell types (epithelial, fibroblast, neuroendocrine, endothelial and immune cells) present in the developing prostate. The CAF and NPF libraries were made from primary cultures of fibroblasts, which are likely heterogeneous populations of fibroblast subgroups. Comparison of the three tag libraries demonstrated that the CAF and EMB libraries shared a greater number of tags with unique UGID (1404 tags; 1342 UGIDs) than the CAF and NPF (851 tags; 777 UGIDs) or the NPF and EMB profiles (1064 tags; 1054 UGIDs). Tags identified as unique or shared between libraries are listed in Supplementary File 1.
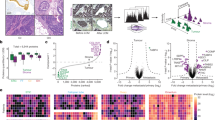
Overlapping and specific Tag expression between CAF, NPF and fetal (EMB) human prostate, and comparison with other data sets. (a) A Venn diagram illustrates the overlap of tags between CAF, NPF and EMB libraries. In all, 12 256 tags were co-expressed between all three libraries, and these comprise ubiquitous or broadly expressed transcripts. The greatest number of co-expressed tags was between CAF and EMB libraries (1404), while the least co-expressed tags were between CAF and NPF (851); NPF and EMB co-expressed 1064 tags. The EMB library contained the highest number of unique tags (19 449) as well as specific tags (4725), which is most likely due to the complex cellular composition of the fetal prostate in comparison with CAF and NPF primary cells. Tag library details are given in Table 1 and Tag lists are in Supplementary File 1. Comparison of the CAF with NPF sample identified CAF-enriched and CAF-depleted Tags. In all, 500 tags with gene symbols were identified as enriched (up) in CAF vs NPF, while 281 tags with gene symbols were depleted (down) in CAF vs NPF. These are listed in Supplementary File 2, and were compared with existing data sets (below). (b) Comparison of CAF-enriched (up) or -depleted (down) transcripts with those identified in CD90+ve stromal cells. Pascal et al. (2009) identified 7379 transcripts enriched in CD90+ve stromal cells and 199 (of 500 CAF enriched) were co-expressed in our data set, while 6298 transcripts were decreased in CD90+ve cells and 105 (of 281 CAF depleted) were co-expressed with our data set. (c) Comparison of CAF-enriched (up) or -depleted (down) transcripts with those identified in reactive stroma. Dakhova et al. (2009) identified 4969 transcripts enriched in reactive stroma and 135 (of 500) CAF-enriched tags were co-expressed in our data set, while 5449 transcripts were decreased in reactive stroma and 94 (of 281) CAF-depleted tags were co-expressed with our data set. Approximately 30–40% of the CAF-enriched or -depleted transcripts were present in either CD90+ve stromal cells or reactive stroma. Fifty-seven transcripts were identified as enriched in CAFs and co-expressed in both CD90+ve stromal cells and reactive stroma, while 29 transcripts were depleted in CAFs and co-expressed in both CD90+ve stromal cells and reactive stroma (Supplementary File 2). Of note, ASPN was identified as CAF enriched and co-expressed in CD90+ve stromal cells and reactive stroma, while NKTR was identified as CAF depleted and decreased in CD90+ve stromal cells and reactive stroma.
Identification of CAF-enriched or -depleted transcripts and comparison with reactive stroma and CD90+ve CAFs
CAFs are involved in regulation of tumourigenesis, and to determine significant changes in tag levels we performed statistical analysis using JumpStart software. CAF and NPF tag lists (cl123) were compared and fold differences calculated (after zeros were converted to ones) followed by statistical evaluation. We identified 671 tags (500 with a gene symbol) as increased (P<0.05) and 356 tags (281 with a gene symbol) as decreased in CAFs vs NPFs (P<0.05). We identified three genes that were present as both up and down in CAFs (ALDH7A1, LYRM2 and MRCL3), due to tag splice variants. To establish that our samples showed previously identified markers associated with CAFs, we interrogated the lists for genes previously studied in prostate cancer stroma. We identified CXCL12, HIF1α, GFRα1, ERα, Hes1 and FIGF (VEGFD) as enriched in CAFs (Allinen et al., 2004; Orimo et al., 2005) and PHB, WFDC1 and CALD as depleted in CAFs (Franco et al., 2011; Kiskowski et al., 2011). Tags with a gene symbol were carried forward for comparison with published microarray data of prostate stroma.
CD90 was reported as enriched in prostate cancer stroma was used for microarray analysis of CD90+ve isolated prostate tumour stromal cells (CD90+ve cells vs CD49a+ve cells, normal fibroblasts; Pascal et al., 2009). In addition, Dakhova et al. (2009) have reported a microarray gene profile of reactive prostate stroma (grade 3) associated with prostate cancer progression and reduced biochemical recurrence-free survival. We compared transcripts enriched or depleted in CAFs with CD90+ve cells and reactive stroma (enriched and depleted). To facilitate comparison of microarray and tag profiles, we compared the data by gene symbol using MS Access. The unique and shared gene expression results are illustrated in Venn diagrams (Figures 1b and c). We identified 57 enriched and 29 depleted transcripts common to CAFs, reactive stroma and CD90+ve stromal cells and their identities are listed in Supplementary File 2. ASPN was identified as CAF enriched and co-expressed in CD90+ve stromal cells and reactive stroma, while NKTR was identified as CAF depleted and decreased in CD90+ve stromal cells and reactive stroma.
Gene Ontology analysis of gene expression data
To identify key processes in CAFs, we used GeneGO software to perform functional classification of well-annotated transcripts with a gene symbol: (1) expressed in both CAF and EMB libraries; (2) enriched/depleted in CAFs vs NPFs; and (3) enriched/depleted in CAFs vs reactive stroma or CD90+ve stromal cells. Gene symbols or Unigene ID was used as gene identifiers. The top 10 enriched process networks and pathway maps are listed in Figure 2 and Supplementary File 3. This analysis co-identified ASPN, FBLN1, FN and OGN as among the top CAF-enriched molecules.
Gene Ontology analysis of enriched and depleted processes and networks in CAFs vs NPFs. GeneGo software was used to compare transcripts identified in CAF with NPF. CAF-enriched processes were predominantly those involved in prostate development, but also included those involved in other reproductive organ growth such as uterus, oocyte and vagina. The processes most depleted in CAFs were associated with cell cycle, suggesting a lower proliferative rate in CAFs. Gene Ontology analysis was also applied to molecules enriched or depleted in CAFs and co-expressed in CD90+ve stromal cells and reactive stroma (Supplementary File 3). The top networks enriched in CAFs included ASPN, FBLN1, FN and OGN.
Selection of candidates for further analysis
We applied several criteria to the selection of candidate molecules for further analysis, which included the following.
We selected secreted and peripheral membrane molecules from the CAF-enriched/depleted molecules (P<0.05) using GeneALaCarte (http://www.genecards.org/).
We selected stromal candidates that showed overlapping gene expression in CAFs vs NPFs (P<0.05) and reactive stroma (Dakhova et al., 2009) and/or CD90+ve stromal cells (Pascal et al., 2009). These were further compared with other gene expression data on prostate stroma and prostate cancer stroma (Madar et al., 2009; Gregg et al., 2010).
Developmental and cancer stroma expressed candidates were identified by comparing gene expression in the CAF and EMB tag profiles and further comparison with developmental prostate mesenchyme (ventral mesenchymal pad, VMP) (Vanpoucke et al., 2007).
Our selected stromal candidates were filtered to identify extracellular protein distribution and stromal localization using the Human Protein Atlas website (http://www.proteinatlas.org/). The analysis identified 15 molecules, 7 that were enriched in CAFs (ASPN, CTSK, FBLN1, FN1, OGN, PARVA and ZEB1) and 5 that were depleted in CAFs (CAV1, CFH, NKTR, S100A6 and STC1) (Table 2). In addition to these 12 molecules, DCN, FHL1 and SPARC were selected from our previous studies of developmental prostatic mesenchyme (Vanpoucke et al., 2007). Table 2 lists the Tag counts of the candidate molecules in the CAF, NPF and EMB libraries, as well as in our previous serial analysis of gene expression (SAGE) profiling of prostatic mesenchyme. This provided a comparison of transcript abundance between adult prostate stroma and developing prostate mesenchyme, as well as between human and rat prostate. Furthermore, co-identification in both sets of libraries (Tag and serial analysis of gene expression) provided support for stromal expression.
QrtPCR validation of selected mRNAs in CAF and NPF patient-matched samples
To validate mRNA expression of the 15 identified candidate molecules in CAFs and to examine whether transcripts were consistently differentially expressed between CAFs and NPFs, QrtPCR was performed on the samples used for Tag profiling plus six additional patient-matched pairs of CAF/NPFs (Figure 3). Human fetal prostate (15–18 weeks) was also included in the analysis to establish mRNA expression levels in development. The CAF/NPFs were functionally tested using previously described methods; CAFs produced tumours when recombined with an epithelial cell line, whereas NPF samples did not (data not shown) (Olumi et al., 1999). In each case, the increase/decrease (P<0.05) of transcript level measured by qrtPCR confirmed the Tag profiling results. QrtPCR analysis of additional CAF/NPF samples showed some variability in candidate gene expression across the sample pairs, which indicates the heterogeneity inherent in patient CAF samples due to variable patient progression and outcome. CXCL12 was increased in 5 of 7 of the CAF samples, consistent with our previous results with prostate CAFs (Ao et al., 2007), and similar to findings in breast cancer stroma (Allinen et al., 2004; Orimo et al., 2005). WFDC1 was decreased in 7 of 7 of the CAF samples, as previously reported (Watson et al., 2004; Supplementary Figure 1). In general, of the Tag identified enriched molecules, CTSK, FN and OGN showed the most consistent pattern of CAF enrichment in CAFs (of the seven selected). For the CAF-depleted molecules, CAV1 S100A6 and STC1 showed the most consistent reduction in CAFs (of the five selected). Taken together, the QrtPCR analysis determined that our candidates were expressed in all of the seven CAF lines examined; however, there appeared to be significant variability in patterns of increased or decreased expression levels reflecting inter-patient differences in rate of disease progression. This may reflect the observation that there are multiple independent mechanisms and pathways involved in CAF pro-tumourigenic signalling, and the role of cellular heterogeneity in CAF function (Franco et al., 2011; Kiskowski et al., 2011).
QrtPCR validation of selected mRNAs in CAF and NPF patient-matched samples and in fetal prostate. QrtPCR for candidate mRNAs was performed on the CAF and NPF samples used for Tag profiling (labelled CAF1), as well as six further matched pairs of functionally tested CAFs and NPFs (labelled 2–7). In all candidates, the Tag count data matched the QrtPCR transcript level and confirmed enrichment or depletion of candidate transcripts in the CAF/NPF sample used for Tag profiling. Of the CAF-enriched molecules: ASPN was increased in two and decreased in three CAF/NPF pairs, CTSK was increased in three CAF/NPF pairs, FBLN1 was increased in three and decreased in two CAF/NPF pairs, FN was increased in five CAF/NPF pairs, OGN was increased in five and decreased in one CAF/NPF pairs, PARVA was increased in two and decreased in four CAF/NPF pairs, ZEB1 was increased in one and decreased in two CAF/NPF pairs. Of the CAF-depleted molecules: CAV1 was decreased in four CAF/NPF pairs, CFH was increased in three and decreased in four CAF/NPF pairs, NKTR was increased in three and decreased in three CAF/NPF pairs, S100A6 was increased in two and decreased in five CAF/NPF pairs, STC1 was increased in one and decreased in four CAF/NPF pairs. As controls, CXCL12 was increased in five and decreased in two CAF/NPF pairs, while WDFC1 was decreased in seven CAF/NPF pairs (Supplementary Figure 1; Supplementary Table 1). In addition, transcript levels were measured in fetal human prostate (EMB) to determine relative mRNA levels, and there was good (but not complete) correlation with candidate Tag counts in the EMB library (Table 2).
Expression of identified candidate proteins in fetal human prostate
To examine the tissue and cellular distribution of our identified candidate molecules in fetal human prostate (18 weeks, n=3), we performed immunohistochemistry (Figure 4). Negative control immunohistochemistry is shown in Supplementary Figure 6. Expression of ASPN, FBLN, PARVA, ZEB1, CAV1, CFH, NKTR and STC1 was observed in a subset of mesenchyme at the outer periphery of the organ. Expression of CTSK and S100A6 was highly restricted to a small subset of cells in the mesenchyme, while OGN was broadly expressed throughout the mesenchyme. CAV1, CTSK, FN, OGN, S100A6 and ZEB1 were localized to the mesenchyme only. The remaining candidates ASPN, CFH, FBLN1, NKTR, PARVA and STC1 exhibited some expression or association with epithelial ducts in addition to their mesenchymal localization. ASPN, SPARC and STC1 are secreted molecules; and therefore, their expression/association with the epithelium may be the result of their secretion from the mesenchyme and binding to the epithelium. Immunolocalization of smooth muscle α-actin (SMACT), myosin (MYO) and E-cadherin (ECAD) were used to identify the mesenchymal and epithelial cell compartments (Supplementary Figure 2). DCN exhibited a localization to mesenchyme in the centre of the fetal prostate, with a peri-urethral distribution, while FHL1 localized to a subset of mesenchyme and smooth muscle, and SPARC showed mesenchymal and epithelial expression (Supplementary Figure 2).
Distribution of identified candidate proteins in fetal human prostate. Samples are orientated with the posterior at the top and the anterior at the bottom, UR denotes urethra, SV denotes seminal vesicles, and selected prostatic ducts are highlighted with arrows in (a, b). The scale bar represents 500 μm. (a–m) Low-magnification images: expression of ASPN, FBLN, PARVA, ZEB1, CAV1, CFH, NKTR and STC1 was observed in a subset of prostatic mesenchyme; and ASPN, FBLN, PARVA, CAV1 and NKTR showed enrichment in prostatic mesenchyme at the periphery of the organ where epithelial branching morphogenesis is concentrated. Expression of CTSK and S100A6 was highly restricted to a small subset of cells in the mesenchyme, while OGN was broadly expressed throughout the mesenchyme. (n–y) High-magnification images: mesenchyme-specific staining for CTSK, FBLN, FN, OGN, PARVA, CAV1, S100A6 and STC1. There was some evidence of nuclear staining in epithelia for ASPN, while ZEB1, CFH, NKTR and STC1 showed limited expression in a subset of epithelia. Scale bar is 50 μm, E, epithelia; M, mesenchyme.
High-power images confirmed the mesenchyme-specific staining for CTSK, FBLN, FN, OGN, PARVA, CAV1, STC1, DCN and FHL1. There was some evidence of nuclear staining in epithelia for ASPN, while ZEB1, CFH, NKTR, STC1 and SPARC showed limited expression in a subset of epithelia (Figure 4; Supplementary Figure 2).
Expression of identified candidate proteins in benign prostate
To determine the immunolocalization of the candidate proteins in benign prostate, we used sections from patient transurethral resection of the prostates (n=3). Figure 5 shows representative areas of benign prostate determined by their tissue histology and immunolocalization of microseminoprotein, beta (MSMB) (Whitaker et al., 2010; Supplementary Figure 5). In areas with normal prostate glands, most of the candidate molecules (CTSK, FBLN1, FN, OGN, PARVA, ZEB1, CAV1, CFH, S100A6 DCN and FHL1) were localized to the stroma only. ASPN, NKTR and SPARC showed some expression/association with ductal epithelia (Figure 5; Supplementary Figure 3). ASPN and STC1 were low or absent from stroma adjacent to benign glands. NKTR is a peripheral membrane protein, while ASPN and SPARC are secreted proteins; therefore, the epithelial staining associated with ASPN and SPARC may be due to stromally expressed protein binding to sites upon epithelial cells. The nuclear epithelial staining observed for ASPN was unexpected, but may represent a novel function for this protein.
Distribution of identified candidate proteins in benign prostate. Low-magnification images (a–m) showed that there was little expression of ASPN, CTSK, FBLN, ZEB1 and STC1 in the stroma, and was limited to few cells or small areas. FN, OGN, PARVA, CAV1 and S100A6 showed broad staining throughout the stroma (scale bar is 200 μm). High-magnification images (n–y) showed that FN, OGN, PARVA, CAV1 and CFH were robustly expressed in most stromal cells, while CTSK, FBLN, NKTR and STC1 were expressed in a small subset of the stroma. (n–y) High-magnification images: most candidates showed stromal expression, CTSK, FBLN, ZEB1, NKTR and STC1, were expressed in few cells and stromal subsets, while FN, OGN, PARVA, CAV1, CFH and S100A6 showed robust stromal staining. ASPN showed some nuclear staining in epithelia, and NKTR showed some surface staining of epithelia (scale bar is 50 μm). Benign prostate was obtained from patients undergoing transurethral resection of the prostate, and was confirmed as non-tumour using microseminoprotein, beta (MSMB) staining (Supplementary Figure 5).
Expression of identified candidate proteins in prostate cancer
Most candidate molecules showed stromal-specific staining in prostate cancer, and a few showed localized regions of more intense staining. ASPN, PARVA, S100A6 and STC1 showed strong staining in stromal subsets, while CTSK, FN, OGN, ZEB1, CAV1, CFH, DCN, FHL1 and SPARC showed expression throughout the stroma, and FBLN and NKTR showed expression in few cells dispersed through the stroma (Figure 6; Supplementary Figure 4). Comparison of candidate expression patterns between benign (Figure 5) and cancer (Figure 6) supported the increase in transcript expression observed for ASPN, CTSK, FBLN and ZEB1, but there was no apparent increase for FN, OGN and PARVA. Similarly, decreased stromal expression in prostate cancer was not apparent for CAV1, CFH, NKTR, S100A6 and STC1, though the immunolocalization data are not quantitative and are correlative in comparison with the QrtPCR data (Figure 3). ASPN, NKTR, S100A6 and ZEB1 expression was also observed in epithelial cells; NKTR is a peripheral membrane protein, ASPN appeared to be nuclear, and both S100A6 and ZEB1 have been reported to be expressed in epithelia. The expression of stromal markers in epithelial cells might suggest a role in EMT, and thus further colocalization experiments were performed. Similarly, there appeared to be expression of ASPN and STC1 in localized areas of stroma associated with tumour epithelia, and these stromal markers were examined for evidence of co-expression in stromal subsets.
Distribution of identified candidate proteins in prostate cancer. (a–m) Low-magnification images (scale bar is 200 μm): all molecules showed stromal expression and ASPN, PARVA, CAV1, S100A6 and STC1 showed a localization to discrete areas of stroma and stromal subsets, while CTSK, FN, OGN, ZEB1 and CFH showed expression throughout the stroma. ASPN and STC1 showed staining in localized stromal areas containing tumour epithelia. (n–y) High-magnification images (scale bar is 50 μm): this confirmed that all candidates showed stromal staining, and also identified some epithelial staining for ASPN, ZEB1, NKTR and S100A6. Nuclear staining in epithelia was observed for ASPN and NKTR, while S100A6 was observed in some epithelia. Prostate cancer tissue was confirmed as tumour tissue by absence of epithelial microseminoprotein, beta (MSMB) expression (Supplementary Figure 5).
Colocalization of identified candidates with E-cadherin to determine co-expression of stromal markers in tumour epithelia
We performed confocal colocalization studies with ASPN, NKTR, S100A6 and E-cadherin to determine whether the stromally expressed molecules were also expressed in epithelia (Figures 7a–c). ASPN, NKTR and S100A6 were expressed in stroma and also showed colocalization with E-cadherin, though in only a small proportion of epithelia. This suggested that they might be markers of EMT, and we performed further colocalization of ASPN, NKTR and S100A6 with both PanCK and ZEB1. PanCK was chosen as our epithelial marker to confirm the epithelial colocalization observed with E-cadherin, while ZEB1 has been shown to mark some EMT cells. We observed colocalization of ASPN, NKTR and S100A6 with ZEB1; however, there was little or no colocalization with PanCK-positive epithelia. To examine whether ASPN and STC1 were expressed in distinct and/or overlapping stromal areas, we performed colocalization studies with these and PanCK. The data showed that there were stromal areas, which expressed either ASPN or STC1 individually, and there were other areas of co-expression. These areas lacked PanCK expression and thus were stromal rather than epithelial.
Colocalization of identified candidates to determine co-expression with epithelial and stromal markers. To examine whether ASPN, NKTR and S100A6 were co-expressed in both stroma and a subset of epithelia, we used confocal colocalization with E-cadherin. (a–c) Overlap between all three molecules and E-cadherin (highlighted by white arrows), in addition to expression in the stroma is shown. Further colocalization of ASPN, NKTR and S100A6 with both ZEB1 and PanCK was performed to confirm the E-cadherin results and to determine whether there was co-expression with ZEB1 in both stroma and epithelia. Stromal colocalization was observed, and there was limited evidence of epithelial colocalization (d–f, white arrows), S100A6 was present in rare basal epithelia as previously observed. To determine whether areas of stroma showed co-expression of ASPN and STC1, these were colocalized (in addition to PanCK to distinguish epithelia (g, h)). There were areas of independent ASPN and STC1 expression (marked with green and red asterisks) as well as areas of co-expression (marked by yellow asterisk).
Discussion
Mesenchyme and stroma have a key role in regulating prostate development and growth, while CAFs in tumour stroma control tumour progression via potent paracrine acting pathways and other mechanisms (Cunha et al., 2002; Bhowmick et al., 2004). We have applied Tag profiling to human CAF, NPF and fetal prostate to identify molecules expressed in prostatic stroma, and focused upon secreted or surface bound molecules present in development and disease. A requirement for a stromal paracrine acting regulator of epithelia is that it is secreted or surface bound, and such molecules would be good therapeutic targets due to their availability and mechanism of action. Our data showed significant overlap with existing data on tumour stroma (Dakhova et al., 2009; Madar et al., 2009; Pascal et al., 2009; Gregg et al., 2010) as well as identifying molecules not previously known to be expressed in mesenchyme and CAFs. We believe that there is significant variability in the cellular composition and differentiation of tumour stroma, and this variability is reflected in transcript profiling data presented here and previously published. CAFs and NPFs are grown as primary cultures of fibroblasts, and their composition is likely to be less heterogeneous than tumour stroma in vivo. The heterogeneity of tumour stroma and CAFs is a key element of their pro-tumourigenic activity (Franco et al., 2011; Kiskowski et al., 2011).
We note that there was most overlap of transcript expression between CAFs and fetal prostate (Figure 1a), and that developmental branching morphogenesis was the top-identified process in CAFs (Figure 2), suggesting strong similarity in pathways expressed in development and disease (Joesting et al., 2005). Such similarities have been noted in profiling studies of whole prostate tumour tissue and developing prostate that focused upon molecules and pathways in tumour epithelia (Schaeffer et al., 2008; Pritchard et al., 2009).
Examination of candidate molecule mRNA levels in paired CAF/NPF samples by QrtPCR showed significant variability between patients (Figure 3; Supplementary Table 1); however, there was general correlation between the Tag library levels and those measured in additional CAF/NPF pairs. We note that there seem to be very few stromal molecules that show uniform increase or decrease in CAFs, for example, CXCL12 or WFDC1. Other data sets support the expression of our identified candidates in prostate stroma (Dakhova et al., 2009; Madar et al., 2009; Pascal et al., 2009; Gregg et al., 2010). The changes in transcript levels that we observed in the Tag libraries and by QrtPCR were supported by the immunohistochemical protein localization, although immunohistochemistry is not quantitative and was performed to establish the tissue localization of candidate proteins rather than their levels. We suggest that the high degree of variability in candidate expression levels in CAFs is partly due to the heterogeneity of CAF cellular composition; there are no reliable markers that uniformly identify CAFs although some of the molecules described here may be useful as markers if further validated. In general, there appears to be a lack of molecules whose expression is restricted to either stroma or CAFs in comparison with numerous molecules expressed in epithelia or both compartments. The identification of molecules expressed only in the stroma or mesenchyme is a prerequisite for defining new EMT markers, and we noted expression of ASPN, NKTR and S100A6 in stroma and epithelia, with evidence of some colocalization with the ZEB1 EMT marker. Further study of these candidates in EMT might be worthwhile. In addition, we noted that ASPN and STC1 marked restricted areas of stroma adjacent to tumour epithelia, and that they showed both distinct and overlapping expression patterns (Figure 7). It is possible that ASPN and STC1 are molecular markers that can be used to identify reactive stroma using immunohistochemistry, though this will require follow-up in large patient cohorts. Reactive stroma is currently defined histologically using Massons trichrome staining, and molecular markers of reactive stroma might be of use for diagnostic purposes given the predictive power of stromal histology (Ayala et al., 2003; Yanagisawa et al., 2007).
Several of molecules that we have identified as expressed in fetal prostate mesenchyme and CAFs have been described in cancer stroma of breast, prostate and other tumours. ASPN is a leucine rich proteoglycan family member that is expressed in breast cancer-associated stroma (Ma et al., 2009), which appears to show some androgen dependence in prostate cancer (Schaeffer et al., 2008) and which binds to TGFβ (Kizawa et al., 2005). CAV1 is downregulated in breast and prostate CAFs and predicts early tumour recurrence and poor outcome (Di Vizio et al., 2009; Witkiewicz et al., 2009). S100A6 is an S100 calcium binding protein family member that is upregulated in many tumours (Lesniak et al., 2009), but is downregulated in prostate cancer via promoter hypermethylation (Rehman et al., 2005). STC1 is pro-survival factor in differentiated cells with variable expression in tumours, which is also involved in developmental mesenchymal–epithelial signalling (Stasko and Wagner, 2001; Joensuu et al., 2008). We suggest that the co-expression of stromal pathways in both mesenchyme and CAFs reflects the similarities between developmental and tumour microenvironment; however, the cellular complexity of the tumour microenvironment may mean that further molecules remain to be identified. The identification of molecules expressed in stromal subsets within the developing fetal prostate is of relevance for understanding potential involvement in CAF identity and pro-tumourigenic signalling because during prostate development there are well-defined subsets of mesenchyme that contain organ-inductive activity which regulates epithelial outgrowth and organogenesis (Timms et al., 1995). The identification of molecules expressed in these key subsets will define markers of these regulatory cell populations as well as paracrine regulators of epithelia made in these cell subgroups (Vanpoucke et al., 2007). Thus, the expression patterns of ASPN, CAV1 and STC1 in restricted areas of developmental prostatic mesenchyme are of particular interest, especially given the restricted expression patterns of ASPN and STC1 in localized areas of tumour stroma and potential as EMT markers. This may indicate that these cell subsets express important regulators of epithelia. We propose that our studies have identified molecules showing stromal restricted expression, some of which are expressed in stromal subsets. These may be used to further characterize the tumour microenvironment and elucidate the cellular heterogeneity.
Materials and methods
Tag profiling
RNA from a patient-matched pair of CAF and NPF cells and pooled embryonic prostate tissue were submitted for Tag profiling by Solexa/Illumina (San Diego, CA, USA) technology (http://www.Illumina.com). Briefly, mRNA was covalently attached to oligo (dT) beads and reverse transcribed. DpnII and MmeI were used as the anchoring and tagging restriction enzymes, respectively. Sequencing was carried out using the Illumina Genome Analyzer. The Illumina tag profiling protocol identifies mRNA transcripts by their unique, positionally known 20 base pair cDNA tag.
Data analysis
Tag profiling data were received from Solexa in the form of tab delimited text files that listed DNA sequences (20 mer) and the counts for individual sequences, which were mapped against the Human Genome (Unigene: HG201). Annotation of the cDNA signature was classed according to their position relative to the polyadenylation signal and the orientation relative to the source mRNA. Signatures classified as classes 1, 2 or 3 were identified as oriented to the forward strand and/or as the 3′ most sequence close to a polyA signal or polyA tail.
The frequency of signature expression was converted to transcripts per million (tpm) and signatures that were present at a frequency of more than three tpm were selected for further analysis. To determine shared and unique expression, the data sets were compared using the signature sequence Microsoft Access. Statistical comparison of gene expression in the full CAF and NPF data sets was performed using JumpStart software (IBM) http://domino.watson.ibm.com/comm/research_projects.nsf/pages/jumpstart.index.html (Stolovitzky et al., 2005). A 95% confidence interval was applied. The assignment of molecular function was performed using GeneGO software (http://www.GeneGo.com).
Analysis of prostate cancer microarray expression data
We obtained expression profile data sets of CD90+ve and CD49a+ve prostate stroma from Pascal et al., (2009) (GEO accession GSE17906) and patient-matched reactive stroma and normal stroma of the prostate from Dakhova et al. (2009) (GEO accession GSE11682—batch corrected data). Raw data values were unlogged and averaged for similar samples. Fold difference in gene expression was calculated for CD90+ve vs CD49a+ve prostate stroma and normal stroma vs reactive stroma. A threshold of ⩾1.25 fold was selected as the cutoff for differential gene expression in analysis of each data set. Data points were removed for poorly annotated genes with only a sequence identifier or without a gene symbol. Comparison of our CAF/NPF tag profiling data sets (P<0.05) with these published microarray data was performed using Microsoft Access using the gene symbol to filter for unique and shared gene expression.
Tissue collection
Human fetal prostate tissue was obtained following medical termination of pregnancy. Consent was obtained in accordance with UK guidelines and the study was approved by the Lothian Research Ethics Committee (08/S1101/1). The bladder and urethra were excised, and the prostate microdissected before RNA isolation. Human adult prostate tissue was obtained from patients undergoing surgery (transurethral resection of the prostate at the Western General Hospital, Edinburgh). Patient consent was obtained before surgery and the study was approved by ethical review (MREC 02/5/63).
Isolation and culture of primary human cells
Matched pairs of CAFs and NPFs were prepared from human prostate tumours as described previously (Olumi et al., 1999).Briefly, benign tissue and tumours were identified using histopathological analysis of stained frozen sections. Five mm3 of tissue fragments immediately adjacent to identified normal tissue and carcinoma was used. Specimens were digested with collagenase and hyaluronidase and placed into culture in RPMI-1640 containing penicillin, streptomycin and Fungizone supplemented with 10% fetal bovine serum. After 10 days of growth, the fibroblastic cells were separated from contaminating epithelial and endothelial cells by differential trypsinization. Subsequent immunocytochemical characterization confirmed their fibroblastic nature. Following outgrowth of stromal cells, the primary cultures were passaged, and the NPF or CAF phenotype was validated according to their tumourigenic activity in tumour reconstitution assays.
RNA extraction and quantitative RT–PCR
Total RNA was extracted using the RNeasy Mini (Qiagen, Crawley, UK) as per the manufacturer's instructions and RNA concentration and purity was measured on the Agilent 2100 Bioanalyser (Agilent Biotechnologies, Santa Clara, CA, USA). Quantitative PCRs were performed on ABI 7500 machine, using the PowerSybr PCR mastermix (ABI, Warrington, UK). Transcript abundance was normalized to TBP expression. The PCR primer sequences are provided below.

Immunohistochemistry
The histology of sections was examined by haematoxylin staining (H&E). For immunostaining, sections were pressure cooked in 10 mM citric acid, pH 6.0, for 5 min. Sections to be stained with 3′ 3-diaminobenzidine (DAB) were treated following a Human Protein Atlas modified protocol (http://www.proteinatlas.org) using the Immpress peroxidase kit (Vector Laboratories, Burlingame, CA, USA) and species appropriate secondary labelled polymer. Antibody concentrations are listed below. Colocalizations were performed with ECAD (BD Biosciences, Oxford, UK) diluted 1:1000 or PCK (Sigma-Aldrich Company Ltd, Dorset, UK) diluted 1:4000 and ZEB1 (Sigma-Aldrich Company, Ltd, Poole, UK) diluted 1:1000. ASPN, NKTR (Sigma) and S100A6 (Thermo Fisher Scientific, Rockford, IL, USA) were diluted 1:50. Antibodies were visualized with species appropriate peroxidase secondary and Tyramide (Perkin-Elmer, Boston, MA, USA) (Zeb1) or species appropriate conjugated avidin-alexafluor 488, 546 or 633 (Molecular Probes Inc., Eugene, OR, USA). Sections were counterstained with DAPI (Sigma-Aldrich Company Ltd) or TOPRO (Molecular Probes Inc.). Images were captured using a Provis microscope (Olympus Optical Co., London, UK) equipped with a DCS330 camera (Eastman Kodak Co., Rochester, NY, USA). Confocal microscopy was performed on a Zeiss LSM 510 Laser scanning microscope (Carl Zeiss Microimaging Inc., Thornwood, NY, USA).

References
Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D et al. (2004). Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6: 17–32.
Ao M, Franco OE, Park D, Raman D, Williams K, Hayward SW . (2007). Cross-talk between paracrine-acting cytokine and chemokine pathways promotes malignancy in benign human prostatic epithelium. Cancer Res 67: 4244–4253.
Augsten M, Hagglof C, Olsson E, Stolz C, Tsagozis P, Levchenko T, Frederick MJ et al. (2009). CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc Natl Acad Sci USA 106: 3414–3419.
Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M et al. (2003). Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res 9: 4792–4801.
Bhowmick NA, Neilson EG, Moses HL . (2004). Stromal fibroblasts in cancer initiation and progression. Nature 432: 332–337.
Cunha GR, Hayward SW, Wang YZ . (2002). Role of stroma in carcinogenesis of the prostate. Differentiation 70: 473–485.
Dakhova O, Ozen M, Creighton CJ, Li R, Ayala G, Rowley D, Ittmann M et al. (2009). Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res 15: 3979–3989.
Di Vizio D, Morello M, Sotgia F, Pestell RG, Freeman MR, Lisanti MP . (2009). An absence of stromal caveolin-1 is associated with advanced prostate cancer, metastatic disease and epithelial Akt activation. Cell Cycle 8: 2420–2424.
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H et al. (2008). Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14: 518–527.
Franco OE, Jiang M, Strand DW, Peacock J, Fernandez S, Jackson II RS, Revelo MP . (2011). Altered TGF-β signaling in a subpopulation of human stromal cells promotes prostatic carcinogenesis. Cancer Res 71: 1272–1281.
Gregg JL, Brown KE, Mintz EM, Piontkivska H, Fraizer GC . (2010). Analysis of gene expression in prostate cancer epithelial and interstitial stromal cells using laser capture microdissection. BMC Cancer 10: 165.
Hayashi N, Cunha GR . (1991). Mesenchyme-induced changes in the neoplastic characteristics of the Dunning prostatic adenocarcinoma. Cancer Res 51: 4924–4930.
Hayward SW, Cunha GR, Dahiya R . (1996). Normal development and carcinogenesis of the prostate. A unifying hypothesis. Ann NY Acad Sci 784: 50–62.
Hayward SW, Wang Y, Cao M, Hom YK, Zhang B, Grossfeld GD, Sudilovsky D . (2001). Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res 61: 8135–8142.
Joensuu K, Heikkila P, Andersson LC . (2008). Tumor dormancy: elevated expression of stanniocalcins in late relapsing breast cancer. Cancer Lett 265: 76–83.
Joesting MS, Perrin S, Elenbaas B, Fawell SE, Rubin JS, Franco OE, Hayward SW . (2005). Identification of SFRP1 as a candidate mediator of stromal-to-epithelial signaling in prostate cancer. Cancer Res 65: 10423–10430.
Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW et al. (2007). Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449: 557–563.
Kiskowski MA, Jackson II RS, Banerjee J, Li X, Kang M, Iturregui JM et al. (2011). Role for stromal heterogeneity in prostate tumorigenesis. Cancer Res 71: 3459–3470.
Kizawa H, Kou I, Iida A, Sudo A, Miyamoto Y, Fukuda A, Mabuchi A . (2005). An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet 37: 138–144.
Lesniak W, Slomnicki LP, Filipek A . (2009). S100A6 - new facts and features. Biochem Biophys Res Commun 390: 1087–1092.
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC . (2009). Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 11: R7.
Madar S, Brosh R, Buganim Y, Ezra O, Goldstein I, Solomon H et al. (2009). Modulated expression of WFDC1 during carcinogenesis and cellular senescence. Carcinogenesis 30: 20–27.
Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR . (1999). Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 59: 5002–5011.
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R . (2005). Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121: 335–348.
Pascal LE, Goo YA, Vencio RZ, Page LS, Chambers AA, Liebeskind ES . (2009). Gene expression down-regulation in CD90+ prostate tumor-associated stromal cells involves potential organ-specific genes. BMC Cancer 9: 317.
Placencio VR, Sharif-Afshar AR, Li X, Huang H, Uwamariya C, Neilson EG et al. (2008). Stromal transforming growth factor-beta signaling mediates prostatic response to androgen ablation by paracrine Wnt activity. Cancer Res 68: 4709–4718.
Pritchard C, Mecham B, Dumpit R, Coleman I, Bhattacharjee M, Chen Q et al. (2009). Conserved gene expression programs integrate mammalian prostate development and tumorigenesis. Cancer Res 69: 1739–1747.
Rehman I, Cross SS, Catto JW, Leiblich A, Mukherjee A, Azzouzi AR et al. (2005). Promoter hyper-methylation of calcium binding proteins S100A6 and S100A2 in human prostate cancer. Prostate 65: 322–330.
Schaeffer EM, Marchionni L, Huang Z, Simons B, Blackman A, Yu W et al. (2008). Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene 27: 7180–7191.
Stasko SE, Wagner GF . (2001). Stanniocalcin gene expression during mouse urogenital development: a possible role in mesenchymal-epithelial signalling. Dev Dyn 220: 49–59.
Stolovitzky GA, Kundaje A, Held GA, Duggar KH, Haudenschild CD, Zhou D et al. (2005). Statistical analysis of MPSS measurements: application to the study of LPS-activated macrophage gene expression. Proc Natl Acad Sci USA 102: 1402–1407.
Sugimoto H, Mundel TM, Kieran MW, Kalluri R . (2006). Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol Ther 5: 1640–1646.
Thalmann GN, Rhee H, Sikes RA, Pathak S, Multani A, Zhau HE, Marshall FF . (2010). Human prostate fibroblasts induce growth and confer castration resistance and metastatic potential in LNCaP Cells. Eur Urol 58: 162–171.
Thomson AA, Cunha GR . (1999). Prostatic growth and development are regulated by FGF10. Development 126: 3693–3701.
Timms BG, Lee CW, Aumuller G, Seitz J . (1995). Instructive induction of prostate growth and differentiation by a defined urogenital sinus mesenchyme. Microsc Res Tech 30: 319–332.
Tuxhorn JA, McAlhany SJ, Dang TD, Ayala GE, Rowley DR . (2002). Stromal cells promote angiogenesis and growth of human prostate tumors in a differential reactive stroma (DRS) xenograft model. Cancer Res 62: 3298–3307.
Vanpoucke G, Orr B, Grace OC, Chan R, Ashley GR, Williams K et al. (2007). Transcriptional profiling of inductive mesenchyme to identify molecules involved in prostate development and disease. Genome Biol 8: R213.
Watson JE, Kamkar S, James K, Kowbel D, Andaya A, Paris PL, Simko J . (2004). Molecular analysis of WFDC1/ps20 gene in prostate cancer. Prostate 61: 192–199.
Whitaker HC, Warren AY, Eeles R, Kote-Jarai Z, Neal DE . (2010). The potential value of microseminoprotein-beta as a prostate cancer biomarker and therapeutic target. Prostate 70: 333–340.
Witkiewicz AK, Casimiro MC, Dasgupta A, Mercier I, Wang C, Bonuccelli G et al. (2009). Towards a new ‘stromal-based’ classification system for human breast cancer prognosis and therapy. Cell Cycle 8: 1654–1658.
Yanagisawa N, Li R, Rowley D, Liu H, Kadmon D, Miles BJ, Wheeler TM, Ayala GE . (2007). Stromogenic prostatic carcinoma pattern (carcinomas with reactive stromal grade 3) in needle biopsies predicts biochemical recurrence-free survival in patients after radical prostatectomy. Hum Pathol 38: 1611–1620.
Zhao H, Ramos CF, Brooks JD, Peehl DM . (2007). Distinctive gene expression of prostatic stromal cells cultured from diseased versus normal tissues. J Cell Physiol 210: 111–121.
Acknowledgements
We would like to thank Ted Pinner for help with the figure preparation, Donald Dunbar for Bio-informatic advice and Sheila Macpherson for help with colocalization. This work was funded by the MRC WBSe 1276.00.003.00004.01 to AAT and NCI CA126505 to SWH. The MRC Human Reproductive Sciences Unit closed on 31 March 2011. AAT was supported by the MRC and The Prostate Cancer Charity.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the oncogene website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Orr, B., Riddick, A., Stewart, G. et al. Identification of stromally expressed molecules in the prostate by tag-profiling of cancer-associated fibroblasts, normal fibroblasts and fetal prostate. Oncogene 31, 1130–1142 (2012). https://doi.org/10.1038/onc.2011.312
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.312
Keywords
This article is cited by
-
Cathepsin K regulates the tumor growth and metastasis by IL-17/CTSK/EMT axis and mediates M2 macrophage polarization in castration-resistant prostate cancer
Cell Death & Disease (2022)
-
Reformation of the chondroitin sulfate glycocalyx enables progression of AR-independent prostate cancer
Nature Communications (2022)
-
Alterations in the methylome of the stromal tumour microenvironment signal the presence and severity of prostate cancer
Clinical Epigenetics (2020)
-
Osteoglycin promotes meningioma development through downregulation of NF2 and activation of mTOR signaling
Cell Communication and Signaling (2017)
-
Identification of genes expressed in a mesenchymal subset regulating prostate organogenesis using tissue and single cell transcriptomics
Scientific Reports (2017)