Abstract
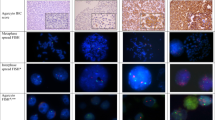
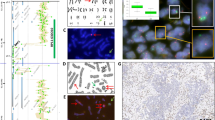
Rearrangements of the neuregulin (NRG1) gene have been implicated in breast carcinoma oncogenesis. To determine the frequency and clinical significance of NRG1 aberrations in clinical breast tumors, a breast cancer tissue microarray was screened for NRG1 aberrations by fluorescent in situ hybridization (FISH) using a two-color split-apart probe combination flanking the NRG1 gene. Rearrangements of NRG1 were identified in 17/382 cases by FISH, and bacterial artificial chromosome array comparative genomic hybridization was applied to five of these cases to further map the chromosome 8p abnormalities. In all five cases, there was a novel amplicon centromeric to NRG1 with a minimum common region of amplification encompassing two genes, SPFH2 and FLJ14299. Subsequent FISH analysis for the novel amplicon revealed that it was present in 63/262 cases. Abnormalities of NRG1 did not correlate with patient outcome, but the novel amplicon was associated with poor prognosis in univariate analysis, and in multivariate analysis was of prognostic significance independent of nodal status, tumor grade, estrogen receptor status, and human epidermal growth factor receptor (HER)2 overexpression. Of the two genes in the novel amplicon, expression of SPFH2 correlated most significantly with amplification. This amplicon may emerge as a result of breakpoints and chromosomal rearrangements within the NRG1 locus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Abbreviations
- TMA:
-
tissue microarray
- FISH:
-
fluorescent in situ hybridization
- HRG:
-
heregulin
- HER:
-
human epidermal growth factor receptor
- IHC:
-
immunohistochemistry
- BAC:
-
bacterial artificial chromosome
- CGH:
-
comparative genomic hybridization
- MFISH:
-
multicolor karyotyping
- MAR:
-
minimum common region of amplification
- BFB:
-
break–fusion–bridge
References
Adelaide J, Chaffanet M, Mozziconacci MJ, Popovici C, Conte N, Fernandez F, Sobol H, Jacquemier J, Pebusque M, Ron D, Lafage-Pochitaloff M and Birnbaum D . (2000). Int. J. Oncol., 16, 683–688.
Adelaide J, Huang HE, Murati A, Alsop AE, Orsetti B, Mozziconacci MJ, Popovici C, Ginestier C, Letessier A, Basset C, Courtay-Cahen C, Jacquemier J, Theillet C, Birnbaum D, Edwards PA and Chaffanet M . (2003). Genes Chromosomes Cancer, 37, 333–345.
Atlas E, Cardillo M, Mehmi I, Zahedkargaran H, Tang C and Lupu R . (2003). Mol. Cancer Res., 1, 165–175.
Bautista S and Theillet C . (1998). Genes Chromosomes Cancer, 22, 268–277.
Bernardino J, Apiou F, Gerbault-Seureau M, Malfoy B and Dutrillaux B . (1998). Genes Chromosomes Cancer, 23, 100–108.
Birnbaum D, Adelaide J, Popovici C, Charafe-Jauffret E, Mozziconacci MJ and Chaffanet M . (2003). Lancet Oncol., 4, 639–642.
Camp RL, Dolled-Filhart M and Rimm DL . (2004). Clin. Cancer Res., 21, 7252–7259.
Coquelle A, Pipiras E, Toledo F, Buttin G and Debatisse M . (1997). Cell, 89, 215–225.
Chi B, DeLeeuw RJ, Coe BP, MacAulay C and Lam WL . (2004). BMC Bioinform., 5, 13.
Chomczynski P and Sacchi N . (1987). Anal. Biochem., 162, 156–159.
Courtay-Cahen C, Morris JS and Edwards PA . (2000). Genomics, 66, 15–25.
Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, Grenier J, Culine S and Theillet C . (2000). Cancer Res., 60, 1077–1083.
de Leeuw RJ, Davies JJ, Rosenwald A, Bebb G, Gascoyne RD, Dyer MJ, Staudt LM, Martinez-Climent JA and Lam WL . (2004). Hum. Mol. Genet., 13, 1827–1837.
Falls DL . (2003). Exp. Cell Res., 284, 14–30.
Huang HE, Chin SF, Ginestier C, Bardou VJ, Adelaide J, Iyer NG, Garcia MJ, Pole JC, Callagy GM, Hewitt SM, Gullick WJ, Jacquemier J, Caldas C, Chaffanet M, Birnbaum D and Edwards PA . (2004). Cancer Res., 64, 6840–6844.
Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C and Lam WL . (2004). Nat. Genet., 36, 299–303.
Jacobson KK, Morrison LE, Henderson BT, Blondin BA, Wilber KA, Legator MS, O'Hare A, Van Stedum SC, Proffitt JH, Seelig SA and Coon JS . (2004). Genes Chromosomes Cancer, 40, 19–31.
Jong K, Marchori E, van der Vaart A, Ylstra B, Weiss M and Meijer G . (2003). Applications of Evolutionary Computing. EvoBIO: Evolutionary Computation and Bioinformatics Lecture Notes in Computer Science, Vol. 2611 Springer: Berlin, pp. 54–65.
Khoury H, Lestou VS, Gascoyne RD, Bruyere H, Li CH, Nantel SH, Dalal BI, Naiman SC and Horsman DE . (2003). Mod. Pathol., 16, 716–724.
Koziczak M, Holbro T and Hynes NE . (2004). Oncogene, 23, 3501–3508.
Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB and van de Rijn M . (2002). Am. J. Pathol., 161, 1557–1565.
Liu X, Baker E, Eyre HJ, Sutherland GR and Zhou M . (1999). Oncogene, 18, 7110–7114.
Makretsov N, Gilks CB, Coldman AJ, Hayes M and Huntsman D . (2003). Hum. Pathol., 34, 1001–1008.
Makretsov N, He M, Hayes M, Chia S, Horsman DE, Sorensen PH and Huntsman DG . (2004a). Genes Chromosomes Cancer, 40, 152–157.
Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MC, Dunn SE, Hayes M, van de Rijn M, Bajdik C and Gilks CB . (2004b). Clin. Cancer Res., 10, 6143–6151.
Nagy P, Vereb G, Sebestyen Z, Horvath G, Lockett SJ, Damjanovich S, Park JW, Jovin TM and Szollosi J . (2002). J. Cell Sci., 115, 4251–4262.
Parker RL, Huntsman DG, Lesack DW, Cupples JB, Grant DR, Akbari M and Gilks CB . (2002). Am. J. Clin. Pathol., 117, 723–728.
Powers CJ, McLeskey SW and Wellstein A . (2000). Endocr. Relat. Cancer, 7, 165–197.
Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA and Ethier SP . (2004). Cancer Res., 64, 40–47.
Rummukainen J, Kytola S, Karhu R, Farnebo F, Larsson C and Isola JJ . (2001). Cancer Genet. Cytogenet., 126, 1–7.
Schaefer G, Fitzpatrick VD and Sliwkowski MX . (1997). Oncogene, 15, 1385–1394.
Sprenger RR, Speijer D, Back JW, De Koster CG, Pannekoek H and Horrevoets AJ . (2004). Electrophoresis, 25, 156–172.
Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C and Sorensen PH . (2002). Cancer Cell, 2, 367–376.
Ugolini F, Adelaide J, Charafe-Jauffret E, Nguyen C, Jacquemier J, Jordan B, Birnbaum D and Pebusque MJ . (1999). Oncogene, 18, 1903–1910.
Wang XZ, Jolicoeur EM, Conte N, Chaffanet M, Zhang Y, Mozziconacci MJ, Feiner H, Birnbaum D, Pebusque MJ and Ron D . (1999). Oncogene, 18, 5718–5721.
Watson SK, deLeeuw RJ, Ishkanian AS, Malloff CA and Lam WL . (2004). BMC Genom., 5, 6.
Wiseman SM, Makretsov N, Neilsen TO, Gilks B, Yorida E, Cheang M, Turbin D, Gelmon K and Huntsman DG . (2005). Cancer, 103, 1770–1777.
Yarden Y and Sliwkowski MX . (2001). Nat. Rev. Mol. Cell. Biol., 2, 127–137.
Acknowledgements
Funding for this project was provided by a Michael Smith Foundation for Health Research (MSFHR) Unit Grant and an Aventis Canada Educational Grant. Torsten O Nielsen and David G Huntsman are MSFHR scholars. Leah M Prentice is an MSFHR trainee.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prentice, L., Shadeo, A., Lestou, V. et al. NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis. Oncogene 24, 7281–7289 (2005). https://doi.org/10.1038/sj.onc.1208892
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208892
Keywords
This article is cited by
-
Fluorescent in situ hybridization has limitations in screening NRG1 gene rearrangements
Diagnostic Pathology (2024)
-
NRG1 fusions in breast cancer
Breast Cancer Research (2021)
-
Genomic characterization of intrinsic and acquired resistance to cetuximab in colorectal cancer patients
Scientific Reports (2019)
-
The Minority Report: Targeting the Rare Oncogenes in NSCLC
Current Treatment Options in Oncology (2014)
-
Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes
Nature Communications (2014)