Abstract
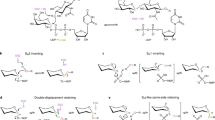
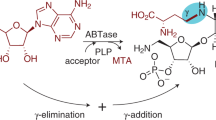
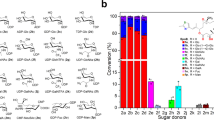
Metabolite glycosylation is affected by three classes of enzymes: nucleotidylyltransferases, which activate sugars as nucleotide diphospho-derivatives, intermediate sugar-modifying enzymes and glycosyltransferases, which transfer the final derivatized activated sugars to aglycon substrates. One of the first crystal structures of an enzyme responsible for the first step in this cascade, α-D-glucopyranosyl phosphate thymidylyltransferase (Ep) from Salmonella, in complex with product (UDP-Glc) and substrate (dTTP) is reported at 2.0 Å and 2.1 Å resolution, respectively. These structures, in conjunction with the kinetic characterization of Ep, clarify the catalytic mechanism of this important enzyme class. Structure-based engineering of Ep produced modified enzymes capable of utilizing 'unnatural' sugar phosphates not accepted by wild type Ep. The demonstrated ability to alter nucleotidylyltransferase specificity by design is an integral component of in vitro glycosylation systems developed for the production of diverse glycorandomized libraries.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Thorson, J.S. et al. Nature's carbohydrate chemists: the enzymatic glycosylation of bioactive bacterial metabolites. Curr. Org. Chem. 5, 89–111 (2001).
Weymouth-Wilson, A.C. The role of carbohydrates in biologically active natural products. Natural Product Reports 14, 99–110 (1997).
Liu, H.-W. & Thorson, J.S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 48, 223–256 (1994).
Kirschning, A., Bechtold, A.F-W. & Rohr, J. Chemical and biochemical aspects of deoxysugars and deoxysugar oligosaccharides. Top. Curr. Chem. 188, 1–84 (1997).
Johnson, D.A. & Liu, H.-W. Mechanisms and pathways from recent deoxysugar biosynthesis research. Curr. Opin. Chem. Biol. 2, 642–649 (1998).
Hallis, T.M. & Liu, H.-W. Learning nature's strategies for making deoxy sugars: pathways, mechanisms, and combinatorial applications. Accts. Chem. Res. 32, 579–588 (1999).
Johnson, D.A. & Liu, H.-W. In Comprehensive chemistry of natural product chemistry. (eds Barton, D., Nakanishi, K. & Meth-Cohn, O.) 311–365 (Elsevier Science, Oxford; 1999).
Trefzer, A., Salas, J. & Bechthold, A. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Natural Product Reports 16, 283–299 (1999).
Bechthold, A. & Rohr, J. In New aspects of bioorganic chemistry. (eds Diederichsen, U., Lindhorst, T.K., Wessjohann, L. & Westerman, B.) 313–348 (Wiley-VCH, Weinheim; 1999).
Madduri, K. et al. Production of the antitumor drug epirubicin (4′-epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nature Biotech. 16, 69–74 (1998).
Hutchinson, C.R. Combinatorial biosynthesis for new drug discovery. Curr. Opin. Microbiol. 1, 319–329 (1998).
Solenberg, P.J. et al. Production of hybrid glycopeptide antibiotics in vitro and in Streptomyces toyocaensis. Chem. Biol. 4, 195–202 (1997).
Lindquist, L., Kaiser, R., Reeves, P.R. & Lindberg, A.A. Purification, characterization and HPLC assay of Salmonella glucose-1-phosphate thymidylyltransferase from the cloned rfbA gene. Eur. J. Biochem. 211, 763–770 (1993).
Jiang, J., Biggins, J.B. & Thorson, J.S. A general enzymatic method for the synthesis of natural and 'unnatural' UDP- and TDP-nucleotide sugars. J. Am. Chem. Soc. 122, 6803–6804 (2000).
Jiang, J., Biggins, J.B. & Thorson, J.S. Expanding the pyrimidine diphosphosugar repertoire: the chemoenzymatic synthesis of amino- and acetamidoglucopyranosyl derivatives. Angew. Chem. 40, 1502–1505 (2001).
Vrielink, A., Ruger, W., Dreissen, H.P.C. & Freemont, P.S. Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 13, 3413–3422 (1994).
Charnock, S.J. & Davies, G.J. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38, 6380–6385 (1999).
Gastinel, L.N. Cambillau, C. & Bourne, Y. Crystal structures of the bovine β4-galatosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 18, 3546–3557 (1999).
Ha, S., Walker, D., Shi, Y. & Walker, S. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9, 1045–1052 (2000).
Brown, K. et al. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridylyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 18, 4096–4107 (1999).
Rossmann, M.G. et al. In The enzymes (ed. I.P.D. Boyyer) 61–102 (Academic Press, New York; 1975).
Branden, C. & Tooze, J. Introduction to protein structure. (Garlan Publishing, Inc., New York; 1991).
Holm, L. & Sander, C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26, 316–319 (1998).
Wedekind, J.P., Frey, P.A. & Rayment, I. Three-dimensional structure of galactose-1-phosphate uridylyltransferase from Escherichia coli at 1.8 Å resolution. Biochemistry 34, 11049–11061 (1995).
Pedersen, L., Benning, M. & Holden, H. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry 34, 13305–13311 (1995).
Kornfeld, S. & Glaser, L. The enzymatic synthesis of thymidine-linked sugars. J. Biol. Chem. 236, 1791–1794 (1961).
Bulik, D.A. et al. UDP-N-acetylglucosamine pyrophosphorylase, a key enzyme in encysting Giardia, is allosterically regulated. J. Biol. Chem. 275, 14722–14728 (2000).
Sheu, K.-F.R., Richard, J.P. & Frey, P.A. Stereochemical courses of nucleotidyltransferase and phosphotransferase action. Uridine diphosphate glucose pyrophosphorylase, galactose-1-phosphate uridylyltransferase, adenylate kinase and nucleoside diphosphate kinase. Biochemistry 18, 5548–5556 (1979).
Segel, I.H. In Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems, (John Wiley & Sons, Inc., New York; 1975).
Volchegursky, Y., Hu, Z., Katz, L. & McDaniel, R. Biosynthesis of the anti-parasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharoropolyspora erythraea. Mol. Microbiol. 37, 752–762 (2000).
Otwinowski, Z. & Minor, W. In Data collection and processing. (eds Sawyer, L., Isaacs, N. & Bailey, S.) 556–562 (SERC Daresbury Laboratory, Warrington, UK; 1993).
CCP4. The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Hendrickson, W.A., Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science, 254, 51–58 (1991).
Miller, R., Gallo, S.M., Khalak, H.G., & Weeks C.M. SnB: crystal structure determination via shake-and-bake. J. Appl. Crystallogr. 32, 120–124 (1994).
Jones, T.A. et al. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. X-PLOR v. 3.1 Manual. (Yale University, New Haven; 1993).
Blankenfeldt, W., Asuncion, M., Lam, J.S. & Naismith, J.H. The structural basis of the catalytic mechanism and regulation of glucose-1-phoshate thymidylyltransferase (Rm1A) EMBO J. 19, 6652–6663 (2000).
Acknowledgements
This contribution was supported in part by the National Institutes of Health to D.B.N. and J.S.T., a Cancer Center Support Grant and a grant from the Special Projects Committee of the Society of Memorial Sloan-Kettering Cancer Center. D.B.N. is a Pew Scholar. J.S.T. is an Alfred P. Sloan Research Fellow and a Rita Allen Foundation Scholar.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Barton, W., Lesniak, J., Biggins, J. et al. Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization. Nat Struct Mol Biol 8, 545–551 (2001). https://doi.org/10.1038/88618
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/88618
This article is cited by
-
Biochemical analysis of leishmanial and human GDP-Mannose Pyrophosphorylases and selection of inhibitors as new leads
Scientific Reports (2017)
-
Cloning and characterization of CalS7 from Micromonospora echinospora sp. calichensis as a glucose-1-phosphate nucleotidyltransferase
Biotechnology Letters (2009)
-
Thermodynamics of binding of divalent magnesium and manganese to uridine phosphates: implications for diabetes-related hypomagnesaemia and carbohydrate biocatalysis
Chemistry Central Journal (2008)
-
Unusual sugar biosynthesis and natural product glycodiversification
Nature (2007)
-
The bittersweet promise of glycobiology
Nature Biotechnology (2001)