Abstract
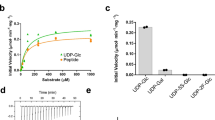
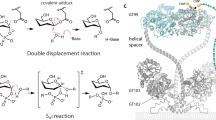
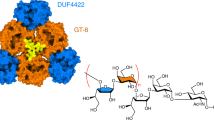
Many bacterial pathogens express lipooligosaccharides that mimic human cell surface glycoconjugates, enabling them to attach to host receptors and to evade the immune response. In Neisseria meningitidis, the galactosyltransferase LgtC catalyzes a key step in the biosynthesis of lipooligosaccharide structure by transferring α-d-galactose from UDP-galactose to a terminal lactose. The product retains the configuration of the donor sugar glycosidic bond; LgtC is thus a retaining glycosyltranferase. We report the 2 Å crystal structures of the complex of LgtC with manganese and UDP 2-deoxy-2-fluoro-galactose (a donor sugar analog) in the presence and absence of the acceptor sugar analog 4′-deoxylactose. The structures, together with results from site-directed mutagenesis and kinetic analysis, give valuable insights into the unique catalytic mechanism and, as the first structure of a glycosyltransferase in complex with both the donor and acceptor sugars, provide a starting point for inhibitor design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Tzeng, Y.L. & Stephens, D.S. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2, 687–700 (2000).
Wakarchuk, W.W., Martin, A., Jennings, M.P., Moxon, E.R. & Richards, J.C. Functional relationships of the genetic locus encoding the glycosyltransferase enzymes involved in expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. J. Biol. Chem. 271, 19166–19173 (1996).
Moran, A.P., Prendergast, M.M. & Appelmelk, B.J. Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease. FEMS Immunol. Med. Microbiol. 16, 105–115 (1996).
Kahler, C. & Stephens, D. Genetic basis for biosynthesis, structure and function of meningococcal lipooligosaccharide (endotoxin). Crit. Rev. Microbiol. 24, 281–334 (1998).
Campbell, J.A., Davies, G.J., Bulone, V. & Henrissat, B. A classification of nucleotide-diphospho-sugar glycosyltransferase based on amino acid sequence similarities. Biochem. J. 329, 929–939 (1997).
Wakarchuk, W., Cunningham, A., Watson, D. & Young, M. Role of paired basic residues in the expression of active recombinant galactosyltransferases from bacterial pathogen Neisseria meningitidis. Protein Eng. 11, 295–302 (1998).
Whitfield, C. & Roberts, I.S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31, 1307–1319 (1999).
Takayama, S. et al. Selective inhibition of β-1,4- and α-1,3-galactosyltransferases: donor sugar-nucleotide based approach. Bioorg. Med. Chem. 7, 401–409 (1999).
Sinnott, M.L. Catalytic mechanisms of enzymatic glycosyl transfer. Chem. Rev. 90, 1171–1202 (1990).
Davies, G., Withers, S.G. & Sinnott, M.L. In Comprehensive biological catalysis, Vol. 1 (ed., Sinnott, M.L.) 119–208 (Academic Press, London; 1997).
Zechel, D.L. & Withers, S.G. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 33, 11–18 (2000).
Vrielink, A., Ruger, W., Driessen, H.P.C. & Freemont, P.S. Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 13, 3413–3422 (1994).
Charnock, S. & Davies, G. Structure of the nucleotide-diphospho-sugar transferase, SpSA from Bacillus subtilin, in native and nucleotide-complexed forms. Biochemistry 38, 6380–6385 (1998).
Gastinel, L., Cambillau, C. & Bourne, Y. Crystal structures of the bovine β4galactosyltransferse catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 18, 3546–3557 (1999).
Ha, S., Walker, D., Shi, Y. & Walker, S. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 9, 1045–1052 (2000).
Pedersen, L.C. et al. Heparan/chondroitin sulfate biosynthesis: structure and mechanism of human glucuronyltransferase I. J. Biol. Chem. 275, 34580–34585 (2000).
Ünligil, U.M. et al. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I; catalytic mechanism and a new protein superfamily. EMBO J. 19, 5269–5280 (2000).
Lu, G. TOP: a new method for protein structure comparison and similarity searches. J. Appl. Crystallogr. 33, 176–183 (2000).
Burkart, M.D. et al. Chemo-enzymatic synthesis of fluorinated sugar nucleotide: useful mechanistic probes for glycosyl transferases. Bioorg. Med. Chem. 8, 1937–1946 (2000).
Thoden, J.B. & Holden, H.M. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 37, 11469–11477 (1998).
Martin, J.L., Johnson, L.N. & Withers, S.G. Comparison of the binding of glucose and glucose 1-phosphate derivatives to T-state glycogen phosphorylase b. Biochemistry 29, 10745–10757 (1990).
Uitdehaag, J.C.M. et al. Catalysis in the α-amylase family: X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase. Nature Struct. Biol. 6, 432–436 (1999).
Brayer, G.D. et al. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry 39, 4778–4791 (2000).
Breton, C., Bettler, E., Joziasse, D.H., Geremia, R.A. & Imberty, A. Sequence-function relationships of prokaryotic and eukaryotic galactosyltransferases. J. Biochem. 123, 1000–1009 (1998).
Kapitonov, D. & Yu, R.K. Conserved domains of glycosyltransferases. Glycobiology 9, 961–978 (1999).
Busch, C. et al. A common motif of eukaryotic glycosyltransferases is essential for the enzymatic activity of large Clostridial cystotoxins. J. Biol. Chem. 273, 19566–19572 (1998).
Shibayamam, K., Ohsuka, S., Toshihiko, T., Yoshickika, A. & Ohta, M. Conserved structural regions involved in the catalytic mechanism of Escherichia coli K-12 WaaO (RfaI). J. Bacteriol. 180, 5313–5318 (1998).
Wiggins, C.A.R. & Munro, S. Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc. Natl. Acad. Sci. USA 95, 7945–7950 (1998).
Hagen, F.K., Hazes, B., Raffo, R., deSa, D. & Tabak, L.A. Structure-function analysis of the UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase. J. Biol. Chem. 274, 6797–6803 (1999).
Moloney, D.J. et al. Fringe is a glycosyltransferase that modifies Notch. Nature 406, 369–375 (2000).
Brückner, K., Perez, L., Clausen, H. & Cohen, S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature 406, 411–415 (2000).
Lougheed, B. M.Sc. thesis. Study of Neisseria meningitidis α-galactosyltransferase. (University of British Columbia; 1998).
Glusker, J. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 42, 1–76 (1991).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Burgi, H., Dunitz, J. & Shefter, E. Nucleophilic addition to a carbonyl group. J. Am. Chem. Soc. 95, 5065–5067 (1973).
Mitchell, E.P. et al. Ternary complex crystal structures of glycogen phosphorylase with the transition state analog nojirimycin tetrazole and phosphate in the T and R states. Biochemistry 35, 7341–7355 (1996).
Watson, K.A. et al. Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question. EMBO J. 18, 4619–4632 (1999).
Artymuik, P.J., Rice, D.W., Poirrette, A.R. & Willett, P. β-Glucosyltransferase and phophorylase reveal their common theme. Nature Struct. Biol. 2, 117–120 (1995).
Holm, L. & Sander, C. Evolutionary link between glycogen phosphorylase and a DNA modifying enzyme. EMBO J. 14, 1287–1293 (1995).
Ly, H.D. & Withers, S.G. Mutagenesis of glycosidases. Annu. Rev. Biochem. 68, 487–522 (1999).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Gosselin, S., Alhussaini, M., Streiff, M.B., Takabayashi, K. & Palcic, M.M. A continuous spectrophotometric assay for glycosyltransferases. Anal. Biochem. 220, 92–97 (1994).
Leatherbarrow, R. Grafit Version 3.0 (Erithacus Software Ltd., Staines, UK; 1990).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Terwilliger, T.C. & Berendzen, J. Automated structure solution for MIR and MAD. Acta Crystallogr. D 55, 849–861 (1999).
Cowtan, K. DM: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994).
McRee, D.E. Practical protein crystallography. A visual protein crystallographic software system for X11/XView. J. Mol. Graphics 10, 44–46 (1992).
Brünger, A. et al. Crystallography and NMR system: a new software suit for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Engh, R. & Huber, R. Accurate bond and angle parameter for X-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 (1991).
Kleywegt, G. & Jones, T. Model building and refinement practice. Methods Enzymol. 277, 208–230 (1997).
Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Kraulis, P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Bacon, D.J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Nicholls, A., Sharp, K. & Honig, B. Protein folding association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296 (1991).
Acknowledgements
We thank H. Bellamy and the staff of the SSRL for access to beamline 1-5 for data collection. We thank D. Dombroski for performing initial crystallization trials on LgtC. Financial support from the Natural Sciences and Engineering Research Council of Canada in terms of a scholarship to H.D.L. and a strategic grant to S.G.W., W.W.W. and N.C.J.S. is gratefully acknowledged. K.P. is a Swedish Research Council for Engineering Sciences Scholar. N.C.J.S. is a MRC Scholar, Burroughs Wellcome New Investigator and Howard Hughes International Scholar.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Persson, K., Ly, H., Dieckelmann, M. et al. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Mol Biol 8, 166–175 (2001). https://doi.org/10.1038/84168
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/84168
This article is cited by
-
Glycosyltransferases as targets for therapeutic intervention in cancer and inflammation: molecular modeling insights
Chemical Papers (2022)
-
Identification and expression analysis of the PtGATL genes under different nitrogen and carbon dioxide treatments in Populus trichocarpa
3 Biotech (2022)
-
A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family
Nature Chemical Biology (2020)
-
A front-face 'SNi synthase' engineered from a retaining 'double-SN2' hydrolase
Nature Chemical Biology (2017)
-
Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT
Scientific Reports (2017)