Abstract
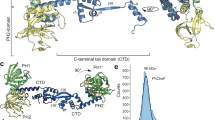
The chemotactic regulator CheY controls the direction of flagellar rotation in Escherichia coli. We have determined the crystal structure of BeF3−-activated CheY from E. coli in complex with an N-terminal peptide derived from its target, FliM. The structure reveals that the first seven residues of the peptide pack against the β4-H4 loop and helix H4 of CheY in an extended conformation, whereas residues 8–15 form two turns of helix and pack against the H4-β5-H5 face. The peptide binds the only region of CheY that undergoes noticeable conformational change upon activation and would most likely be sandwiched between activated CheY and the remainder of FliM to reverse the direction of flagellar rotation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Egger, L.A., Park, H. & Inouye, M. Genes To Cells 2, 167–184 (1997).
Parkinson, J.S. & Kofoid, E.C. Annu. Rev. Genet. 26, 71–112 (1992).
Falke, J.J., Bass, R.B., Butler, S.L., Chervitz, S.A. & Danielson, M.A. Annu. Rev. Cell Dev. Biol. 13, 457–512 (1997).
Djordjevic, S. & Stock, A.M. J. Struct. Biol. 124, 189–200 (1998).
Rombel, I., North, A., Hwang, I., Wyman, C. & Kustu, S. Cold Spring Harbor Symp. Quant. Biol. 63, 157–166 (1998).
Kustu, S., North, A.K. & Weiss, D.S. Trends Biochem. Sci. 16, 397–402 (1991).
Roman, S.J., Meyers, M., Volz, K. & Matsumura, P. J. Bacteriol. 174, 6247–6255 (1992).
Sockett, H., Yamaguchi, S., Kihara, M., Irikura, V.M. & Macnab, R.M. J. Bacteriol. 174, 793–806 (1992).
Barak, R. & Eisenbach, M. Curr. Top. Cell Regul. 34, 137–158 (1996).
Toker, A.S. & Macnab, R.M. J. Mol. Biol. 273, 623–634 (1997).
Bren, A. & Eisenbach, M. J. Mol. Biol. 278, 507–514 (1998).
Welch, M., Oosawa, K., Aizawa, S.-I. & Eisenbach, M. Biochemistry 33, 10470–10476 (1994).
McEvoy, M.M., Bren, A., Eisenbach, M. & Dahlquist, F.W. J. Mol. Biol. 289, 1423–1433 (1999).
Yan, D. et al. Proc. Natl. Acad. Sci. USA 96, 14789–14794 (1999).
Cho, H.S. et al. J. Mol. Biol. 297, 543–551 (2000).
Lewis, R.J., Brannigan, J.A., Muchova, K., Barak, I. & Wilkinson, A.J. J. Mol. Biol. 294, 9–15 (1999).
Birck, C. et al. Structure 7, 1505–1515 (1999).
Matthews, M.A.A., Tang, H.L. & Blair, D.F. J. Bacteriol. 180, 5580–5590 (1998).
Toker, A.S., Kihara, M. & Macnab, R.M. J. Bacteriol. 178, 7069–7079 (1996).
Halkides, C.J. et al. Biochemistry 39, 5280–5286 (2000).
Lo Conte, L., Chothia, C. & Janin, J. J. Mol. Biol. 285, 2177–2198 (1999).
Aurora, R. & Rose, G.D. Protein Sci. 7, 21–38 (1998).
Shuster, M., Zhao, R., Bourret, R.B. & Collins, E.J. J. Biol. Chem. 275, 19752–19758 (2000).
Kern, D. et al. Nature 402, 894–898 (1999).
Lee, J., Owens, J.T., Hwang, I., Meares, C. & Kustu, S. J. Bacteriol. 182, 5188–5192 (2000).
Lee, J. Ph.D. Thesis. Phosphorylation Induced Interdomain Communication in the Response Regulator of NtrC from Salmonella typhimurium. (University of California; 2000).
Shi, W., Yang, Z., Geng, Y., Wolinsky, L.E. & Lovett, M.A. J. Bacteriol. 180, 231–235 (1998).
Canale-Parola, E. Free-living Saccharolytic Spirochetes, the genus Spirochaeta. In The prokaryotes, Vol. 4 (ed. Balows, A.) 329–333 (Springer-Verlag, New York; 1992).
Brünger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Bailey, S. Acta Crystallogr. D 50, 760–763 (1994).
Kleywegt, G.J. & Jones, T.A. Acta Crystallogr. D 55, 941–944 (1999).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Nicholls, A., Sharp, K. & Honig, B. Proteins 11, 281–296 (1991).
Volz, K. & Matsumura, P. J. Biol. Chem. 266, 15511–15519 (1993).
Acknowledgements
We thank H. Bellamy for performing the X-ray fluorescence scan and help in designing the MAD experiment at SSRL. We thank T. Earnest (ALS) and D. Shin (Physical Biosciences Division, LBL) for helpful advice and encouragement. This work was supported by the Office of Energy Research, Office of Health and Environmental Research, Health Effects Research Division of the U.S. Department of Energy (D.E.W.) and National Institutes of Health (S.K.) and through instrumentation grants from the U.S. Department of Energy and the National Science Foundation (D.E.W.). This work was done (partially) at SSRL which is operated by the Department of Energy, Office of Basic Energy Sciences. The SSRL Biotechnology Program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and by the Department of Energy, Office of Biological and Environmental Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, SY., Cho, H., Pelton, J. et al. Crystal structure of an activated response regulator bound to its target. Nat Struct Mol Biol 8, 52–56 (2001). https://doi.org/10.1038/83053
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/83053
This article is cited by
-
Mutations in the stator protein PomA affect switching of rotational direction in bacterial flagellar motor
Scientific Reports (2022)
-
Effect of Heterologous Expression of Chemotaxis Proteins from Genus Thermotoga on the Growth Kinetics of Escherichia coli Cells
Bulletin of Experimental Biology and Medicine (2019)
-
Chemical Approaches to Studying Labile Amino Acid Phosphorylation
Topics in Current Chemistry (2017)
-
Focus on phosphoaspartate and phosphoglutamate
Amino Acids (2011)
-
Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus
The EMBO Journal (2005)