Abstract
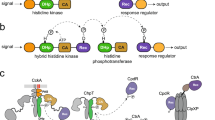
Histidine protein kinases and response regulators form the basis of phosphotransfer signal transduction pathways. Commonly referred to as two-component systems, these modular and adaptable signaling schemes are prevalent in prokaryotes. Structures of the core domains of histidine kinases reveal a protein kinase fold different from that of the Ser/Thr/Tyr protein kinase family, but similar to that of other ATP binding domains. Recent structure determinations of phosphorylated response regulator domains indicate a conserved mechanism for the propagated conformational change that accompanies phosphorylation of an active site Asp residue. The altered molecular surface promotes specific protein–protein interactions that mediate the downstream response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Stock, A., Koshland, D.E., Jr. & Stock, J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc. Natl. Acad. Sci. USA 82, 7989–7993 (1985).
Nixon, B.T., Ronson, C.W. & Ausubel, F.M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83, 7850–7854 (1986).
Stock, J.B., Surette, M.G., Levit, M. & Park, P. In Two-component signal transduction (eds, Hoch, J.A. & Silhavy, T.J.), 25–51 (American Society for Microbiology, Washington, DC; 1995).
Stock, A.M., Robinson, V.L. & Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000).
Mizuno, T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4, 161–168 (1997).
Throup, J.P. et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35, 566–576 (2000).
Fabret, C., Feher, V.A. & Hoch, J.A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181, 1975–1983 (1999).
Smith, D.R. et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179, 7135–7155 (1997).
Mizuno, T. His-Asp phosphotransfer signal transduction. J. Biochem. 123, 555–563 (1998).
Chang, C. & Stewart, R.C. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117, 723–731 (1998).
Perraud, A.-L., Weiss, V. & Gross, R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 7, 115–120 (1999).
Falke, J.J., Bass, R.B., Butler, S.L., Chervitz, S.A. & Danielson, M.A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13, 457–512 (1997).
Maeda, T., Wurgler-Murphy, S.M. & Saito, H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369, 242–245 (1994).
Thomason, P.A. et al. An intersection of the cAMP/PKA and two-component signal transduction systems in Dictyostelium. EMBO J. 17, 2838–2845 (1998).
Bilwes, A.M., Alex, L.A., Crane, B.R. & Simon, M.I. Structure of CheA, a signal-transducing histidine kinase. Cell 96, 131–141 (1999).
Tomomori, C. et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nature Struct. Biol. 6, 729–734 (1999).
Tanaka, T. et al. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature 396, 88–92 (1998).
Birck, C. et al. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure Fold. Des. 7, 1505–1515 (1999).
Kern, D. et al. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 402, 894–898 (1999).
Lewis, R.J., Brannigan, J.A., Muchová, K., Barák, I. & Wilkinson, A.J. Phosphorylated aspartate in the structure of a response regulator protein. J. Mol. Biol. 294, 9–15 (1999).
Cho, H.S. et al. NMR structure of activated CheY. J. Mol. Biol. 297, 543–551 (2000).
Halkides, C.J. et al. The 1.9 Å resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry 39, 5280–5286 (2000).
Stock, J.B., Stock, A.M. & Mottonen, J.M. Signal transduction in bacteria. Nature 344, 395–400 (1990).
Wigley, D.B., Davies, G.J., Dodson, E.J., Maxwell, A. & Dodson, G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 (1991).
Prodromou, C. et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90, 65–75 (1997).
Ban, C. & Yang, W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell 95, 541–552 (1998).
Mushegian, A.R., Bassett, D.E., Jr., Boguski, M.S., Bork, P. & Koonin, E.V. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc. Natl. Acad. Sci. USA 94, 5831–5836 (1997).
Ban, C., Junop, M. & Yang, W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97, 85–97 (1999).
Brino, L. et al. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275, 9468–9475 (2000).
Yeh, K.-C., Wu, S.-H., Murphy, J.T. & Lagarias, J.C. A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508 (1997).
Yeh, K.-C. & Lagarias, J.C. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95, 13976–13981 (1998).
Elich, T.D. & Chory, J. Phytochrome: If it looks and smells like a histidine-kinase, is it a histidine kinase? Cell 91, 713–716 (1997).
Wu, J., Ohta, N., Zhao, J.L. & Newton, A. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96, 13068–13073 (1999).
Ryazanov, A.G., Pavur, K.S. & Dorovkov, M.V. Alpha-kinases: a new class of protein kinases with a novel catalytic domain. Curr. Biol. 9, R43–R45 (1999).
Varughese, K.I., Madhusudan, Zhou, X.Z., Whiteley, J.M. & Hoch, J.A. Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase. Mol. Cell. 2, 485–493 (1998).
Zhou, H., Lowry, D.F., Swanson, R.V., Simon, M.I. & Dahlquist, F.W. NMR studies of the phosphotransfer domain of the histidine kinase CheA from Escherichia coli: assignments, secondary structure, general fold, and backbone dynamics. Biochemistry 34, 13858–13870 (1995).
Kato, M., Mizuno, T., Shimizu, T. & Hakoshima, T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell 88, 717–723 (1997).
Xu, Q. & West, A.H. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J. Mol. Biol. 292, 1039–1050 (1999).
Park, H. & Inouye, M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J. Bacteriol. 179, 4382–4390 (1997).
Singh, M., Berger, B., Kim, P.S., Berger, J.M. & Cochran, A.G. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl. Acad. Sci. USA 95, 2738–2743 (1998).
Aravind, L. & Ponting, C.P. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176, 111–116 (1999).
Taylor, B.L. & Zhulin, I.B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506 (1999).
Gong, W. et al. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95, 15177–15182 (1998).
Pellequer, J.L., Wager-Smith, K.A., Kay, S.A. & Getzoff, E.D. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA 95, 5884–5890 (1998).
Aravind, L. & Ponting, C.P. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22, 458–459 (1997).
Artymiuk, P.J., Rice, D.W., Mitchell, E.M. & Willet, P. Structural resemblance between the families of bacterial signal-transducing proteins and of G proteins revealed by graph theoretical techniques. Protein Eng. 4, 39–43 (1990).
Chen, J.M., Lee, G., Murphy, R.B., Brandt-Rauf, P.W. & Pincus, M.R. Comparisons between the three-dimensional structures of the chemotactic protein CheY and the normal Gly 12‐p21 protein. Int. J. Pept. Protein Res. 36, 1–6 (1990).
Stock, J.B., Lukat, G.S. & Stock, A.M. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu. Rev. Biophys. Biophys. Chem. 20, 109–136 (1991).
Jencks, W.P. The utilization of binding energy in coupled vectorial processes. Adv. Enzymol. 51, 75–106 (1980).
Tanford, C. Twenty questions concerning the reaction cycle of the sarcoplasmic reticulum calcium pump. CRC Crit. Rev. Biochem. 17, 123–151 (1984).
Volz, K. Structural conservation in the CheY superfamily. Biochemistry 32, 11741–11753 (1993).
Stock, A.M., Mottonen, J.M., Stock, J.B. & Schutt, C.E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature 337, 745–749 (1989).
Djordjevic, S., Goudreau, P.N., Xu, Q., Stock, A.M. & West, A.H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95, 1381–1386 (1998).
Muller-Dieckmann, H.J., Grantz, A.A. & Kim, S.H. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure Fold. Des. 7, 1547–1556 (1999).
Gouet, P. et al. Structural transitions in the FixJ receiver domain. Structure 7, 1517–1526 (1999).
Baikalov, I. et al. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry 37, 3665–3676 (1998).
Volkman, B.F., Nohaile, M.J., Amy, N.K., Kustu, S. & Wemmer, D.E. Three-dimensional solution structure of the N-terminal receiver domain of NtrC. Biochemistry 34, 1413–1424 (1995).
Solà, M., Gomis-Rüth, F.X., Serrano, L., González, A. & Coll, M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 285, 675–687 (1999).
Madhusudan et al. Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis. Structure 4, 679–690 (1996).
Lukat, G.S., McCleary, W.R., Stock, A.M. & Stock, J.B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89, 718–722 (1992).
Lukat, G.S., Stock, A.M. & Stock, J.B. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry 29, 5436–5442 (1990).
Martinez-Hackert, E. & Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269, 301–312 (1997).
Mizuno, T. & Tanaka, I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 24, 665–667 (1997).
Osuna, J., Soberon, X. & Morett, E. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 6, 543–555 (1997).
Pelton, J.G., Kustu, S. & Wemmer, D.E. Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol. 292, 1095–1110 (1999).
Zhu, X., Rebello, J., Matsumura, P. & Volz, K. Crystal structures of CheY mutants Y106W and T87I/Y106W: CheY activation correlates with movement of residue 106. J. Biol. Chem. 272, 5000–5006 (1997).
Nohaile, M., Kern, D., Wemmer, D., Stedman, K. & Kustu, S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J. Mol. Biol. 273, 299–316 (1997).
Yan, D. et al. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA 96, 14789–14794 (1999).
Lewis, R.J. et al. Domain swapping in the sporulation response regulator Spo0A. J. Mol. Biol. 297, 757–770 (2000).
Ames, S.K., Frankema, N. & Kenney, L.J. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc. Natl. Acad. Sci. USA 96, 11792–11797 (1999).
Hwang, I., Thorgeirsson, T., Lee, J., Kustu, S. & Shin, Y.K. Physical evidence for a phosphorylation-dependent conformational change in the enhancer-binding protein NtrC. Proc. Natl. Acad. Sci. USA 96, 4880–4885 (1999).
Anand, G.A., Goudreau, P.N., Lewis, J.K. & Stock, A.M. Evidence for phosphorylation-dependent conformational changes in methylesterase CheB. Protein Sci. 9, 898–906 (2000).
Welch, M., Chinardet, N., Mourey, L., Birck, C. & Samama, J.-P. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nature Struct. Biol. 5, 25–29 (1998).
McEvoy, M.M., Hausrath, A.C., Randolph, G.B., Remington, S.J. & Dahlquist, F.W. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc. Natl. Acad. Sci. USA 95, 7333–7338 (1998).
McEvoy, M.M., Bren, A., Eisenbach, M. & Dahlquist, F.W. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J. Mol. Biol. 289, 1423–1433 (1999).
Acknowledgements
We thank G. Anand, K. Gunsalus, J. Hurley and C. Waldburger for comments on the manuscript and/or for providing unpublished information, and the NIH and HHMI for support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Robinson, V., Buckler, D. & Stock, A. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Mol Biol 7, 626–633 (2000). https://doi.org/10.1038/77915
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/77915
This article is cited by
-
Evidence of Cross-Regulation in Two Closely Related Pyruvate-Sensing Systems in Uropathogenic Escherichia coli
The Journal of Membrane Biology (2018)
-
The yeasts phosphorelay systems: a comparative view
World Journal of Microbiology and Biotechnology (2017)
-
Computational studies on histidine kinase protein BaeS to target multidrug-resistant Salmonella
Medicinal Chemistry Research (2013)
-
Molecular cloning, sequence analysis and structure modeling of OmpR, the response regulator of Aeromonas hydrophila
Molecular Biology Reports (2012)
-
Identification of ColR binding consensus and prediction of regulon of ColRS two-component system
BMC Molecular Biology (2009)