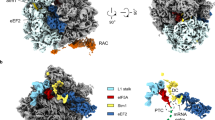
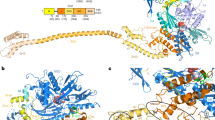
Abstract
Chaperones, which assist protein folding are essential components of every living cell. The yeast ribosome-associated complex (RAC) is a chaperone that is highly conserved in eukaryotic cells. The RAC consists of the J protein Zuo1 and the unconventional Hsp70 homolog Ssz1. The RAC heterodimer stimulates the ATPase activity of the ribosome-bound Hsp70 homolog Ssb, which interacts with nascent polypeptide chains to facilitate de novo protein folding. In addition, the RAC–Ssb system is required to maintain the fidelity of protein translation. Recent work reveals important details of the unique structures of RAC and Ssb and identifies how the chaperones interact with the ribosome. The new findings start to uncover how the exceptional chaperone triad cooperates in protein folding and maintenance of translational fidelity and its connection to extraribosomal functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hallstrom, T.C., Katzmann, D.J., Torres, R.J., Sharp, W.J. & Moye-Rowley, W.S. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 1147–1155 (1998).This work revealed, for the first time, that Ssz1 is an Hsp70 homolog that is involved in the activation of the transcription factor Pdr1.
Zhang, S., Lockshin, C., Herbert, A., Winter, E. & Rich, A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 11, 3787–3796 (1992).
Boyer, A.S., Grgurevic, S., Cazaux, C. & Hoffmann, J.S. The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. J. Mol. Biol. 425, 4767–4781 (2013).
Yan, W. et al. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17, 4809–4817 (1998).This study revealed that the J-domain protein Zuo1 binds to ribosomes via ribosomal RNA and functions in concert with the Hsp70 Ssb.
Nelson, R.J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M. & Craig, E.A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105 (1992).This work revealed that a large fraction of the Hsp70 Ssb is ribosome associated and involved in translation.
Mayer, M.P. & Bukau, B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 (2005).
Craig, E.A., Huang, P., Aron, R. & Andrew, A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156, 1–21 (2006).
Kampinga, H.H. & Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010).
Gautschi, M. et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98, 3762–3767 (2001).The study revealed that the J-domain protein Zuo1 forms a stable complex with the Hsp70 Ssz1, termed ribosome-associated complex (RAC).
Gautschi, M., Mun, A., Ross, S. & Rospert, S. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99, 4209–4214 (2002).
Hundley, H. et al. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA 99, 4203–4208 (2002).The work in refs. 10,11 revealed that Zuo1, Ssz1, and Ssb form a ribosome-bound chaperone triad.
Otto, H. et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA 102, 10064–10069 (2005).This work identified mammalian RAC consisting of the J-domain protein ZRF1/MPP1 and the Hsp70 Hsp70L1.
Pfund, C., Huang, P., Lopez-Hoyo, N. & Craig, E.A. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12, 3773–3782 (2001).
Peisker, K., Chiabudini, M. & Rospert, S. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803, 662–672 (2010).
Mayer, M.P. & Kityk, R. Insights into the molecular mechanism of allostery in Hsp70s. Front Mol Biosci 2, 58 (2015).
Huang, P., Gautschi, M., Walter, W., Rospert, S. & Craig, E.A. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12, 497–504 (2005).This group demonstrated that RAC can stimulate the ATPase activity of Ssb and revealed that Ssz1 function does not require ATP hydrolysis.
Conz, C. et al. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J. Biol. Chem. 282, 33977–33984 (2007).
Leidig, C. et al. Structural characterization of a eukaryotic chaperone—the ribosome-associated complex. Nat. Struct. Mol. Biol. 20, 23–28 (2013).
Weyer, F.A., Gumiero, A., Gesé, G.V., Lapouge, K. & Sinning, I. Structural insights into a unique Hsp70-Hsp40 interaction in the eukaryotic ribosome-associated complex. Nat. Struct. Mol. Biol. 24, 144–151 (2017).In this work, the authors solved the crystal structure of full-length Ssz1 in a complex with Zuo1 ND, providing the first structural insight into the unique complex.
Jaiswal, H. et al. The chaperone network connected to human ribosome-associated complex. Mol. Cell. Biol. 31, 1160–1173 (2011).This study revealed that mRAC cooperates with an Hsp70 partner, which does not directly interact with the ribosome.
Sahi, C. & Craig, E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 104, 7163–7168 (2007).
Chen, D.H., Huang, Y., Liu, C., Ruan, Y. & Shen, W.H. Functional conservation and divergence of J-domain-containing ZUO1/ZRF orthologs throughout evolution. Planta 239, 1159–1173 (2014).
Fiaux, J. et al. Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J. Biol. Chem. 285, 3227–3234 (2010).
Zhang, Y. et al. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat. Struct. Mol. Biol. 21, 1042–1046 (2014).In this work, the authors discovered that Zuo1 spans both ribosomal subunits.
Kaschner, L.A., Sharma, R., Shrestha, O.K., Meyer, A.E. & Craig, E.A. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim. Biophys. Acta 1853, 1035–1045 (2015).
Lee, K., Sharma, R., Shrestha, O.K., Bingman, C.A. & Craig, E.A. Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits. Nat. Struct. Mol. Biol. 23, 1003–1010 (2016).In this paper, the authors identified and characterized the critical contacts between Zuo1 and ES12 of the 40S and H24 of the 60S ribosomal subunits.
Gumiero, A. et al. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nat. Commun. 7, 13563 (2016).This group solved the crystal structure of full-length ATP-bound Ssb and, together with work from ref. 55, identified Ssb's ribosome-binding domain.
Kityk, R., Kopp, J., Sinning, I. & Mayer, M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 (2012).
Kityk, R., Vogel, M., Schlecht, R., Bukau, B. & Mayer, M.P. Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308 (2015).
Halic, M. et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 (2004).
Rospert, S. Ribosome function: governing the fate of a nascent polypeptide. Curr. Biol. 14, R386–R388 (2004).
Halic, M. & Beckmann, R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 15, 116–125 (2005).
Grudnik, P., Bange, G. & Sinning, I. Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782 (2009).
Nyathi, Y., Wilkinson, B.M. & Pool, M.R. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2392–2402 (2013).
Bhushan, S. et al. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 (2010).
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 (2011).
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M., Arpagaus, S. & Ban, N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 (2011).
Lin, P.J., Jongsma, C.G., Pool, M.R. & Johnson, A.E. Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J. Cell Biol. 195, 55–70 (2011).
Zhang, Y., Wölfle, T. & Rospert, S. Interaction of nascent chains with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 of Saccharomyces cerevisiae. J. Biol. Chem. 288, 33697–33707 (2013).
Peisker, K. et al. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19, 5279–5288 (2008).
Rakwalska, M. & Rospert, S. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 9186–9197 (2004).This study revealed, for the first time, that RAC and Ssb are required for the maintenance of translational fidelity.
Rospert, S., Rakwalska, M. & Dubaquié, Y. Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev. Physiol. Biochem. Pharmacol. 155, 1–30 (2005).
Hatin, I., Fabret, C., Namy, O., Decatur, W.A. & Rousset, J.P. Fine-tuning of translation termination efficiency in Saccharomyces cerevisiae involves two factors in close proximity to the exit tunnel of the ribosome. Genetics 177, 1527–1537 (2007).
Muldoon-Jacobs, K.L. & Dinman, J.D. Specific effects of ribosome-tethered molecular chaperones on programmed -1 ribosomal frameshifting. Eukaryot. Cell 5, 762–770 (2006).
Chiabudini, M., Conz, C., Reckmann, F. & Rospert, S. Ribosome-associated complex and Ssb is required for translational repression induced by polylysine segments within nascent chains. Mol. Cell. Biol. 32, 4769–4779 (2012).
Chiabudini, M. et al. Release factor eRF3 mediates premature translation termination on polylysine-stalled ribosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 34, 4062–4076 (2014).
Kim, S.Y. & Craig, E.A. Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone ssb or zuo1 to cations, including aminoglycosides. Eukaryot. Cell 4, 82–89 (2005).
Albanèse, V., Yam, A.Y., Baughman, J., Parnot, C. & Frydman, J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124, 75–88 (2006).
Verghese, J., Abrams, J., Wang, Y. & Morano, K.A. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 76, 115–158 (2012).
Ogle, J.M., Carter, A.P. & Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28, 259–266 (2003).
Garreau de Loubresse, N. et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522 (2014).
Zaher, H.S. & Green, R. Fidelity at the molecular level: lessons from protein synthesis. Cell 136, 746–762 (2009).
Velichutina, I.V., Hong, J.Y., Mesecar, A.D., Chernoff, Y.O. & Liebman, S.W. Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18 S rRNA. J. Mol. Biol. 305, 715–727 (2001).
Tselika, S., Konstantinidis, T.C. & Synetos, D. Two nucleotide substitutions in the A-site of yeast 18S rRNA affect translation and differentiate the interaction of ribosomes with aminoglycoside antibiotics. Biochimie 90, 908–917 (2008).
Hanebuth, M.A. et al. Multivalent contacts of the Hsp70 Ssb contribute to its architecture on ribosomes and nascent chain interaction. Nat. Commun. 7, 13695 (2016).
Pfund, C. et al. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17, 3981–3989 (1998).This study demonstrated for the first time that ribosome-bound Ssb directly interacts with nascent chains.
Gautschi, M. et al. The yeast N(α)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23, 7403–7414 (2003).
Willmund, F. et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 (2013).This work identified cotranslational substrates of Ssb and demonstrated widespread aggregation of newly synthesized proteins in cells lacking Ssb.
Pechmann, S., Willmund, F. & Frydman, J. The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 (2013).
Gong, Y. et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 5, 275 (2009).
Yam, A.Y., Albanèse, V., Lin, H.T. & Frydman, J. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280, 41252–41261 (2005).The studies in refs. 61,75–77 revealed that the Hsp110 homolog Sse1 interacts with, and acts as an NEF, for Ssa or Ssb.
Meyer, A.E., Hung, N.J., Yang, P., Johnson, A.W. & Craig, E.A. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 104, 1558–1563 (2007).
Greber, B.J., Boehringer, D., Montellese, C. & Ban, N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1228–1233 (2012).
Chernoff, Y.O., Newnam, G.P., Kumar, J., Allen, K. & Zink, A.D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19, 8103–8112 (1999).
Kiktev, D.A., Melomed, M.M., Lu, C.D., Newnam, G.P. & Chernoff, Y.O. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 96, 621–632 (2015).
Chernoff, Y.O. & Kiktev, D.A. Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 62, 677–685 (2016).
von Plehwe, U. et al. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 23, 2102–2115 (2009).
Koplin, A. et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68 (2010).In refs. 68 and 69, the authors demonstrated that RAC and Ssb serve a specific function in ribosome biogenesis.
Albanèse, V., Reissmann, S. & Frydman, J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J. Cell Biol. 189, 69–81 (2010).
Preissler, S. & Deuerling, E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 (2012).
Shulga, N., James, P., Craig, E.A. & Goldfarb, D.S. A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J. Biol. Chem. 274, 16501–16507 (1999).
Prunuske, A.J., Waltner, J.K., Kuhn, P., Gu, B. & Craig, E.A. Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proc. Natl. Acad. Sci. USA 109, 472–477 (2012).This work revealed that not only Ssz1 but also Zuo1 can activate the transcription factor Pdr1.
Ducett, J.K. et al. Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. J. Mol. Biol. 425, 19–31 (2013).This work revealed that the unfolding of the Zuo1 4HB leads to ribosome release and activation of the transcription factor Pdr1 via direct interaction.
Hübscher, V. et al. The Hsp70 homolog Ssb and the 14-3-3 protein Bmh1 jointly regulate transcription of glucose repressed genes in Saccharomyces cerevisiae. Nucleic Acids Res. 44, 5629–5645 (2016).This work revealed that Ssb and Bmh1 are involved in the post-translational regulation of the signaling kinase SNF1.
Dragovic, Z., Broadley, S.A., Shomura, Y., Bracher, A. & Hartl, F.U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 (2006).
Shaner, L., Wegele, H., Buchner, J. & Morano, K.A. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280, 41262–41269 (2005).
Raviol, H., Sadlish, H., Rodriguez, F., Mayer, M.P. & Bukau, B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 (2006).
Abrams, J.L., Verghese, J., Gibney, P.A. & Morano, K.A. Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. J. Biol. Chem. 289, 13155–13167 (2014).
Kakiuchi, K. et al. Proteomic analysis of in vivo 14-3-3 interactions in the yeast Saccharomyces cerevisiae. Biochemistry 46, 7781–7792 (2007).
Panni, S. et al. Combining peptide recognition specificity and context information for the prediction of the 14-3-3-mediated interactome in S. cerevisiae and H. sapiens. Proteomics 11, 128–143 (2011).
Obsil, T. & Obsilova, V. Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672 (2011).
Coblitz, B., Wu, M., Shikano, S. & Li, M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 580, 1531–1535 (2006).
Dombek, K.M., Kacherovsky, N. & Young, E.T. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279, 39165–39174 (2004).This work provided the first evidence for a role of Ssb in glucose repression.
Hübscher, V., Mudholkar, K. & Rospert, S. The yeast Hsp70 homolog Ssb: a chaperone for general de novo protein folding and a nanny for specific intrinsically disordered protein domains. Curr. Genet. 63, 9–13 (2017).
Broach, J.R. Nutritional control of growth and development in yeast. Genetics 192, 73–105 (2012).
van Heusden, G.P. & Steensma, H.Y. Yeast 14-3-3 proteins. Yeast 23, 159–171 (2006).
Kleppe, R., Martinez, A., Døskeland, S.O. & Haavik, J. The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell Dev. Biol. 22, 713–719 (2011).
Bustos, D.M. The role of protein disorder in the 14-3-3 interaction network. Mol. Biosyst. 8, 178–184 (2012).
Sluchanko, N.N. & Gusev, N.B. Moonlighting chaperone-like activity of the universal regulatory 14–3-3 proteins. FEBS J. 284, 1279–1295 (2017).
Smock, R.G. & Gierasch, L.M. Sending signals dynamically. Science 324, 198–203 (2009).
Mashaghi, A. et al. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 (2016).
Liu, Q. & Craig, E.A. Molecular biology: mature proteins braced by a chaperone. Nature 539, 361–362 (2016).
Paul, S. & Moye-Rowley, W.S. Multidrug resistance in fungi: regulation of transporter-encoding gene expression. Front. Physiol. 5, 143 (2014).
Eisenman, H.C. & Craig, E.A. Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol. Microbiol. 53, 335–344 (2004).
Boyer, L.A., Latek, R.R. & Peterson, C.L. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163 (2004).
Hundley, H.A., Walter, W., Bairstow, S. & Craig, E.A. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science 308, 1032–1034 (2005).The studies in refs. 12 and 96 revealed that ZRF1/MPP11 interacts with ribosomes and is the human homolog of yeast Zuo1.
Gracheva, E. et al. ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J. Cell Biol. 213, 185–200 (2016).
Richly, H. et al. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468, 1124–1128 (2010).In this work, authors discovered that ZRF1/MPP11 and Zuo1 can bind monoubiquitin.
Aloia, L., Gutierrez, A., Caballero, J.M. & Di Croce, L. Direct interaction between Id1 and Zrf1 controls neural differentiation of embryonic stem cells. EMBO Rep. 16, 63–70 (2015).
Rath, S.K. et al. Silencing of ZRF1 impedes survival of estrogen receptor positive MCF-7 cells and potentiates the effect of curcumin. Tumour Biol. 37, 12535–12546 (2016).
Resto, V.A. et al. A putative oncogenic role for MPP11 in head and neck squamous cell cancer. Cancer Res. 60, 5529–5535 (2000).
Papadopoulou, T. & Richly, H. On-site remodeling at chromatin: how multiprotein complexes are rebuilt during DNA repair and transcriptional activation. BioEssays 38, 1130–1140 (2016).
Boyer, L.A. et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10, 935–942 (2002).
Richly, H. & Di Croce, L. The flip side of the coin: role of ZRF1 and histone H2A ubiquitination in transcriptional activation. Cell Cycle 10, 745–750 (2011).
Tovchigrechko, A. & Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 (2006).
Raue, U., Oellerer, S. & Rospert, S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282, 7809–7816 (2007).
Ashe, M.P., De Long, S.K. & Sachs, A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833–848 (2000).
Swaney, D.L. et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 (2013).
Weinert, B.T. et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 (2013).
Albuquerque, C.P. et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 (2008).
Ohno, A. et al. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure 13, 521–532 (2005).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through FOR967 (to I.S. and S.R.), GRK1188, SFB1036 and the Leibniz program (to I.S.), SFB 746 (to S.R.), and RO 1028/5-1 (to S.R.), and by the Excellence Initiative of the German federal and state governments [BIOSS-2] (to S.R). I.S. is an investigator of the Cluster of Excellence: CellNetworks. We thank C. Conz, F. Weyer, K. Lapouge, and G. Valentin Gese for careful reading and suggestions on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Y., Sinning, I. & Rospert, S. Two chaperones locked in an embrace: structure and function of the ribosome-associated complex RAC. Nat Struct Mol Biol 24, 611–619 (2017). https://doi.org/10.1038/nsmb.3435
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3435
This article is cited by
-
Structural inventory of cotranslational protein folding by the eukaryotic RAC complex
Nature Structural & Molecular Biology (2023)
-
Structural remodeling of ribosome associated Hsp40-Hsp70 chaperones during co-translational folding
Nature Communications (2022)
-
Deciphering molecular details of the RAC–ribosome interaction by EPR spectroscopy
Scientific Reports (2021)
-
Pathway of Hsp70 interactions at the ribosome
Nature Communications (2021)
-
Innate immunity to prions: anti-prion systems turn a tsunami of prions into a slow drip
Current Genetics (2021)