Abstract
We report the cryo-EM structure of the human 26S proteasome at an average resolution of 3.5 Å, allowing atomic modeling of 28 subunits in the core particle (CP) and 18 subunits in the regulatory particle (RP). The C-terminal residues of Rpt3 and Rpt5 subunits in the RP can be seen inserted into surface pockets formed between adjacent α subunits in the CP. Each of the six Rpt subunits contains a bound nucleotide, and the central gate of the CP α-ring is closed despite RP association. The six pore 1 loops in the Rpt ring are arranged similarly to a spiral staircase along the axial channel of substrate transport, which is constricted by the pore 2 loops. We also determined the cryo-EM structure of the human proteasome bound to the deubiquitinating enzyme USP14 at 4.35-Å resolution. Together, our structures provide a framework for mechanistic understanding of eukaryotic proteasome function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
04 August 2016
In the version of this article initially published online, the section on 'Cryo-EM sample preparation and data acquisition' in the Online Methods contained errors. The units for defocus values were incorrectly reported as millimeters. The correct unit should be micrometers. Images were recorded automatically instead of manually as initially stated. This section of the Online Methods has been rewritten to improve clarity, and these errors have been corrected for the print, PDF and HTML versions of this article.
References
Finley, D., Chen, X. & Walters, K.J. Gates, channels, and switches: elements of the proteasome machine. Trends Biochem. Sci. 41, 77–93 (2016).
Deveraux, Q., Ustrell, V., Pickart, C. & Rechsteiner, M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269, 7059–7061 (1994).
Husnjak, K. et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008).
Zhang, F. et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 (2009).
Wang, F. et al. Structure and mechanism of the hexameric MecA–ClpC molecular machine. Nature 471, 331–335 (2011).
Lander, G.C. et al. Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 (2012).
Yao, T. & Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Verma, R. et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615 (2002).
Nyquist, K. & Martin, A. Marching to the beat of the ring: polypeptide translocation by AAA+ proteases. Trends Biochem. Sci. 39, 53–60 (2014).
Pick, E., Hofmann, K. & Glickman, M.H. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell 35, 260–264 (2009).
Maytal-Kivity, V., Reis, N., Hofmann, K. & Glickman, M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3, 28 (2002).
Estrin, E., Lopez-Blanco, J.R., Chacón, P. & Martin, A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure 21, 1624–1635 (2013).
Tomko, R.J. Jr. et al. A single a helix drives extensive remodeling of the proteasome lid and completion of regulatory particle assembly. Cell 163, 432–444 (2015).
Wang, Q., Young, P. & Walters, K.J. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J. Mol. Biol. 348, 727–739 (2005).
Zhang, N. et al. Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol. Cell 35, 280–290 (2009).
Sakata, E. et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 109, 1479–1484 (2012).
Amerik, A.Y. & Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695, 189–207 (2004).
Lee, B.H. et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532, 398–401 (2016).
Hu, M. et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 (2002).
Hu, M. et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756 (2005).
Löwe, J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268, 533–539 (1995).
Groll, M. et al. Structure of 20S proteasome from yeast at 2.4Å resolution. Nature 386, 463–471 (1997).
Unno, M. et al. The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure 10, 609–618 (2002).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Park, S. et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature 497, 512–516 (2013).
Campbell, M.G., Veesler, D., Cheng, A., Potter, C.S. & Carragher, B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. eLife 4, e06380 (2015).
da Fonseca, P.C. & Morris, E.P. Cryo-EM reveals the conformation of a substrate analogue in the human 20S proteasome core. Nat. Commun. 6, 7573 (2015).
Harshbarger, W., Miller, C., Diedrich, C. & Sacchettini, J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure 23, 418–424 (2015).
Lasker, K. et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA 109, 1380–1387 (2012).
Beck, F. et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. USA 109, 14870–14875 (2012).
da Fonseca, P.C., He, J. & Morris, E.P. Molecular model of the human 26S proteasome. Mol. Cell 46, 54–66 (2012).
Matyskiela, M.E., Lander, G.C. & Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 (2013).
Śledź, P. et al. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. USA 110, 7264–7269 (2013).
Unverdorben, P. et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 111, 5544–5549 (2014).
Aufderheide, A. et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. USA 112, 8626–8631 (2015).
Bohn, S. et al. Localization of the regulatory particle subunit Sem1 in the 26S proteasome. Biochem. Biophys. Res. Commun. 435, 250–254 (2013).
Bashore, C. et al. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat. Struct. Mol. Biol. 22, 712–719 (2015).
Luan, B. et al. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc. Natl. Acad. Sci. USA 113, 2642–2647 (2016).
Lee, B.H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010).
Wang, X. et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553–3565 (2007).
Thompson, D., Hakala, K. & DeMartino, G.N. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J. Biol. Chem. 284, 24891–24903 (2009).
Yu, Y. et al. Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 29, 692–702 (2010).
Tian, G. et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 18, 1259–1267 (2011).
Förster, A., Masters, E.I., Whitby, F.G., Robinson, H. & Hill, C.P. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 18, 589–599 (2005).
Kusmierczyk, A.R., Kunjappu, M.J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 (2008).
Wendler, P., Ciniawsky, S., Kock, M. & Kube, S. Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823, 2–14 (2012).
Banerjee, S. et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351, 871–875 (2016).
Wang, J. et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9, 177–184 (2001).
Yamada-Inagawa, T., Okuno, T., Karata, K., Yamanaka, K. & Ogura, T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J. Biol. Chem. 278, 50182–50187 (2003).
Schweitzer, A. et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc. Natl. Acad. Sci. USA 113, 7816–7821 (2016).
Dambacher, C.M., Worden, E.J., Herzik, M.A., Martin, A. & Lander, G.C. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife 5, e13027 (2016).
Smith, D.M. et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 (2007).
Glynn, S.E., Martin, A., Nager, A.R., Baker, T.A. & Sauer, R.T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009).
Pettersen, E.F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Leggett, D.S., Glickman, M.H. & Finley, D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 (2005).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zhou, Q. et al. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res. 25, 551–560 (2015).
Kucukelbir, A., Sigworth, F.J. & Tagare, H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008).
Tomko, R.J. Jr. & Hochstrasser, M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol. Cell 53, 433–443 (2014).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Acknowledgements
We thank the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for providing facility support. The computation was completed on the 'Explorer 100' cluster system of the Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the National Natural Science Foundation of China 31130002, 31321062, and 31430020 to Y.S.
Author information
Authors and Affiliations
Contributions
X.H., B.L., and Y.S. designed the experiments; X.H. and B.L. expressed and purified human proteasomes and performed biochemical experiments; X.H. and B.L. collected the EM data; X.H., B.L., J.W., and Y.S. performed EM data processing and analysis; X.H., B.L., J.W., and Y.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Purification, characterization and cryo-EM analysis of the human 26S proteasome.
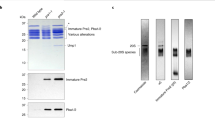
(a) A schematic diagram of the affinity purification protocol for endogenous human proteasome from a modified HEK293T cell line. (b) The purified human proteasome is reasonably homogeneous on native PAGE and exhibits proteolytic activity. The purified human proteasome appeared mainly as a dominant band of a 2:1 complex between the RP and the CP on native PAGE (lane 1). Incubation of the native PAGE gel with the fluorogenic substrate Suc-LLVY-AMC revealed substrate degradation as detected by fluorescence emission of the free AMC moiety (lane 2). (c) Identification of the Rpn and Rpt subunits on SDS-PAGE. The purified human proteasome was applied to SDS-PAGE followed by Coomassie staining (lane 1). The molecular weight markers are shown in lane 2. (d) Results of mass spectrometric (MS) identification of the human proteasomal subunits. All subunits were identified by MS. (e) A representative cryo-EM micrograph of the human proteasome. The proteasome particles are clearly seen in the original micrograph. Scale bar, 50 nm. (f) Representative 2D class averages of the human proteasome. Due to its elongated shape, the proteasome displays a clear preference of orientation on the carbon-coated grid.
Supplementary Figure 2 Flowchart for the cryo-EM analysis of the 26S human proteasome.
(a) 443,359 particles were subjected to 2D classification and subsequent 3D refinement. After 3D classification, 165,669 particles were picked for the following 3D refinement and particle polishing. With C1 and C2 symmetry, reconstructions at an average resolution of 3.8 Å and 3.5 Å were produced respectively. Please refer to Materials and Methods for details. (b) Particle segmentation and re-extraction were performed to improve the quality of the local density maps for the RP. Cryo-EM maps of RP in two conformations were produced, both at 4.6 Å. Please refer to Materials and Methods for details. Structural images in this figure, along with all images involving cryo-EM density maps, were prepared using CHIMERA.
Supplementary Figure 3 Cryo-EM analysis of the 26S human proteasome.
(a) Cryo-EM density maps of the human proteasome calculated with the C1 symmetry have an averaged resolution of 3.8 Å. Two perpendicular views are shown. The resolution range is color-coded. (b) Cryo-EM density maps of the RP calculated with a soft mask on the RP have an averaged resolution of 4.3 Å. (c) FSC curves of the three cryo-EM reconstructions: entire human proteasome with C2 symmetry (magenta), entire human proteasome with C1 symmetry (cyan), and the RP alone (green). (d) Angular distribution for the final reconstruction of the human proteasome with C2 symmetry. Each cylinder represents one view and the height of the cylinder is proportional to the number of particles for that view. The orientation preference is clearly seen. (e) FSC curves of the final refined model versus the overall 3.5 Å map (C2 symmetry) it was refined against (black); of the model refined in the first of the two independent maps used for the gold-standard FSC versus that same map (red); and of the model refined in the first of the two independent maps versus the second independent map (green). The small difference between the red and green curves indicates that the refinement of the atomic coordinates did not suffer from severe overfitting.
Supplementary Figure 4 Cryo-EM density maps of individual subunits in the proteasome.
(a) Overall cryo-EM density maps of the α-ring in the CP calculated with C2 symmetry. Most notably, the central gate for substrate entry is closed, as evidenced by the relatively well-defined density maps at the gate. (b) Overall cryo-EM density maps of the β-ring in the CP calculated with C2 symmetry. The proteolytic chamber is empty. Representative cryo-EM density maps for the seven subunits α1 through α7 are displayed in panels c through i, respectively. Representative density maps for the seven subunits β1 through β7 are displayed in panels j through p, respectively. The side chain features are clearly identifiable in the density maps. (q) Cryo-EM density maps for the nine Rpn subunits in the lid (Rpn3, Rpn4, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn11, Rpn12, and Rpn15). For each of these Rpn subunits, the overall density maps are shown in the left, and the local maps for select secondary structural elements are shown in the right. (r) Cryo-EM density maps for the six Rpt subunits in the base (Rpt1 through Rpt6). For each of these Rpt subunits, the overall density maps are shown in the left, and the local maps for select secondary structural elements are shown in the right. The large and small AAA domains in each Rpt subunit are identified by magenta and green circles, respectively. (s) Cryo-EM density maps for three Rpn subunits in the base (Rpn1, Rpn2, and Rpn10).
Supplementary Figure 5 Cryo-EM density maps of select subcomplexes in the human proteasome.
(a) Cryo-EM density maps of the overall organization in the proteasome. Three perpendicular views are shown. The 18 subunits in the RP are color-coded. (b) Cryo-EM density maps of the lid. The atomic models are shown on the right. In each case, two perpendicular views are shown. The solenoid folds of six Rpn subunits (Rpn3, Rpn5, Rpn6, Rpn7, Rpn9, and Rpn12) radiate from the center. (c) Cryo-EM density maps of the hexameric Rpt ring. The atomic models are shown on the right. In each case, two perpendicular views are shown. The coiled-coil of Rpt6-Rpt3 exhibits excellent density maps, in part because it is stabilized through direct interactions with Rpn3 of the Lid and Rpn2 of the base. In contrast, the other two coiled-coils, formed by Rpt4-5 and Rpt2-1, point into solvent and have discontinuous density maps, likely reflecting their dynamic conformations. (d) Cryo-EM density maps of the heptameric α-ring in two perpendicular views. The atomic model is shown in the right. (e) Cryo-EM density maps of the heptameric β-ring in two perpendicular views. The atomic model is shown in the right.
Supplementary Figure 6 Interactions between the Rpt ring and the CP, and structural comparison between the AAA motor protein p97 and the Rpt subunits.
(a) An overall view on the interface between the Rpt ring and the CP, centered on Rpt1 (left panel). The carboxyl-terminal sequences of Rpt1 run parallel with the surface of the α-ring, and the carboxyl-terminus is not inserted into the α-pocket between subunits α4 and α5 (right panel). (b) An overall view on the interface between the Rpt ring and the CP, centered on Rpt4 (left panel). The carboxyl-terminal sequences of Rpt4 run parallel with the surface of the α-ring, and the carboxyl-terminus is not inserted into the α-pocket between subunits α7 and α1 (right panel). (c) An overall view on the interface between the Rpt ring and the CP, centered on Rpt2 (left panel), and a close-up view on the carboxyl-terminus of Rpt2 (right panel). The carboxyl-terminal nine residues of Rpt2 exhibit no apparent EM density and are left out in the final atomic model. (d) An overall view on the interface between the Rpt ring and the CP, centered on Rpt6 (left panel), and a close-up view on the carboxyl-terminus of Rpt6 (right panel). The carboxyl-terminal 11 residues of Rpt6 exhibit no apparent EM density and are left out in the final atomic model. (e) Structural comparison between p97 and each of the six Rpt subunits with alignment of their large AAA domains. Structures of p97 bound to ADP (PDB accession code 5FTM) and ATP-γS (PDB accession code 5FTN) are colored green and magenta, respectively. This comparison reveals that the conformations of Rpt1, Rpt3, Rpt4, and Rpt5 more closely resemble that of the ATP-γS-bound p97. The conformations of Rpt2 and Rpt6 are closer to that of the ADP-bound p97. (f) Structural comparison between p97 and each of the six Rpt subunits with alignment of both large and small domains. Compared to panel a, the conclusions remain unchanged for all but one Rpt subunit. Rpt3 shows a better overall alignment to the ADP-bound p97.
Supplementary Figure 7 Substrate-translocation channel and the pore loops.
(a) An overall view of the axial channel for substrate translocation. The surface view (left) and a cut-open section at the center of the CP plus Rpt ring (right) are shown. The axial channel (identified by two red lines) in the Rpt ring contains two constriction points, one at the OB ring and the other at the AAA ring. (b) The pore-1 loops from the six Rpt subunits form a spiral staircase along the axial channel. The overall view is shown in the left panel, whereas two perpendicular views on the EM density of the pore-1 loops are shown in the middle and right panels. (c) The pore-2 loops from five Rpt subunits are placed along the axial channel. The overall view is shown in the left panel, whereas two perpendicular views on the EM density of five pore-2 loops are shown in the middle and right panels. The pore-2 loop of Rpt6 exhibits no clear density and was not modeled. (d) The OB domains of the six Rpt subunits assemble into a circular structure. The central constriction point in the OB ring measures approximately 12-18 Å in diameter, insufficient for a folded globular protein to come through. The L23 and L45 loops line the constriction. The cryo-EM density maps are displayed in colored mesh for the six OB domains. Structural features of the inter-subunit interactions in the OB ring are shown below. Similar to the case of the coiled-coils, the six OB domains form three pairs of hetero-dimers. Notably, within each OB hetero-dimer, there is a pair of charge-stabilized H-bonds between an Arg in one Rpt subunit and a Glu in the adjacent Rpt.
Supplementary Figure 8 USP14 binds to the 26S proteasome.
(a) Native PAGE assay of the USP14-proteasome interactions. The purified human proteasome (1.6 picomoles per reaction) was incubated for 20 minutes at 30°C with 16 picomoles of GST-USP14 (USP14G), untagged USP14, or GST. 64 picomoles UbAl were added where indicated. Samples were resolved on a 4% native PAGE gel and visualized by Suc-LLVY-AMC in-gel hydrolysis (upper panel). Binding of USP14G to the proteasome results in marked reduction of its electrophoretic mobility. The bottom panel shows Coomassie blue staining of the gel. Yeast CP was used as a marker. (b) A pull-down assay was performed in the presence and absence of UbAl. The presence of UbAl appears to enhance the interaction of USP14 with the proteasome. The binding of USP14G to proteasome is more obvious than that for untagged USP14.
Supplementary Figure 9 Cryo-EM analysis of the human 26S proteasome bound to USP14–UbAl.
(a) A flow-chart for the cryo-EM analysis of the human 26S proteasome bound to USP14-UbAl. 534,096 particles were subjected to 2D classification and subsequent auto-refinement. After 3D classification, 182,079 particles were picked for the following 3D refinement. With the soft mask on CP-RP2 and RP, reconstructions at average resolutions of 4.2 Å and 4.35 Å, respectively, were produced. Please refer to Materials and Methods for details. (b) Cryo-EM density map of the human 26S proteasome bound to USP14-UbAl calculated with the RP mask displays an average resolution of 4.35 Å. Two perpendicular views are shown. The resolution range is color-coded. Red circles indicate the location of USP14-UbAl. (c) FSC curves of the two cryo-EM reconstructions: human 26S proteasome with the CP-RP2 mask (cyan) and with the RP mask (purple).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Table 1 (PDF 11107 kb)
Rights and permissions
About this article
Cite this article
Huang, X., Luan, B., Wu, J. et al. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 23, 778–785 (2016). https://doi.org/10.1038/nsmb.3273
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3273
This article is cited by
-
Diminished ovarian reserve causes adverse ART outcomes attributed to effects on oxygen metabolism function in cumulus cells
BMC Genomics (2023)
-
POH1/Rpn11/PSMD14: a journey from basic research in fission yeast to a prognostic marker and a druggable target in cancer cells
British Journal of Cancer (2022)
-
Discovery proteomics defines androgen-regulated glycoprotein networks in prostate cancer cells, as well as putative biomarkers of prostatic diseases
Scientific Reports (2021)
-
Proteasome regulation by reversible tyrosine phosphorylation at the membrane
Oncogene (2021)
-
The Hsp70–Hsp90 go-between Hop/Stip1/Sti1 is a proteostatic switch and may be a drug target in cancer and neurodegeneration
Cellular and Molecular Life Sciences (2021)