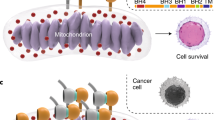
Abstract
MCL-1 is an antiapoptotic BCL-2 family protein that has emerged as a major pathogenic factor in human cancer. Like BCL-2, MCL-1 bears a surface groove whose function is to sequester the BH3 killer domains of proapoptotic BCL-2 family members, a mechanism harnessed by cancer cells to establish formidable apoptotic blockades. Although drugging the BH3-binding groove has been achieved for BCL-2, translating this approach to MCL-1 has been challenging. Here, we report an alternative mechanism for MCL-1 inhibition by small-molecule covalent modification of C286 at a new interaction site distant from the BH3-binding groove. Our structure–function analyses revealed that the BH3 binding capacity of MCL-1 and its suppression of BAX are impaired by molecular engagement, a phenomenon recapitulated by C286W mutagenic mimicry in vitro and in mouse cells. Thus, we characterize an allosteric mechanism for disrupting the antiapoptotic BH3 binding activity of MCL-1, informing a new strategy for disarming MCL-1 in cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Walensky, L.D. & Gavathiotis, E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem. Sci. 36, 642–652 (2011).
Sattler, M. et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997).
Souers, A.J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
van Delft, M.F. et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 (2006).
Deng, J. et al. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell 12, 171–185 (2007).
Konopleva, M. et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10, 375–388 (2006).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Bruncko, M. et al. Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity. J. Med. Chem. 58, 2180–2194 (2015).
Leverson, J.D. et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis. 6, e1590 (2015).
Burke, J.P. et al. Discovery of tricyclic indoles that potently inhibit Mcl-1 using fragment-based methods and structure-based design. J. Med. Chem. 58, 3794–3805 (2015).
Friberg, A. et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J. Med. Chem. 56, 15–30 (2013).
Stewart, M.L., Fire, E., Keating, A.E. & Walensky, L.D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 6, 595–601 (2010).
Cohen, N.A. et al. A competitive stapled peptide screen identifies a selective small molecule that overcomes MCL-1-dependent leukemia cell survival. Chem. Biol. 19, 1175–1186 (2012).
Baell, J. & Walters, M.A. Chemistry: chemical con artists foil drug discovery. Nature 513, 481–483 (2014).
Tan, L. et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc. Natl. Acad. Sci. USA 111, E4869–E4877 (2014).
Zhou, W. et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 462, 1070–1074 (2009).
Follis, A.V. et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat. Chem. Biol. 9, 163–168 (2013).
Gavathiotis, E., Reyna, D.E., Davis, M.L., Bird, G.H. & Walensky, L.D. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol. Cell 40, 481–492 (2010).
Engen, J.R. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 81, 7870–7875 (2009).
Walensky, L.D. et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004).
Laiken, S.L., Printz, M.P. & Craig, L.C. Tritium-hydrogen exchange studies of protein models. I. Gramicidin S-A. Biochemistry 8, 519–526 (1969).
Printz, M.P., Williams, H.P. & Craig, L.C. Evidence for the presence of hydrogen-bonded secondary structure in angiotensin II in aqueous solution. Proc. Natl. Acad. Sci. USA 69, 378–382 (1972).
Shi, X.E. et al. Hydrogen exchange-mass spectrometry measures stapled peptide conformational dynamics and predicts pharmacokinetic properties. Anal. Chem. 85, 11185–11188 (2013).
Endo, A., Sumi, D., Iwamoto, N. & Kumagai, Y. Inhibition of DNA binding activity of cAMP response element-binding protein by 1,2-naphthoquinone through chemical modification of Cys-286. Chem. Biol. Interact. 192, 272–277 (2011).
Ostrem, J.M., Peters, U., Sos, M.L., Wells, J.A. & Shokat, K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013).
Stewart, D.P. et al. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol. Cell. Biol. 30, 3099–3110 (2010).
Lee, E.F. et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J. Biol. Chem. 284, 30508–30517 (2009).
Verdine, G.L. & Walensky, L.D. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin. Cancer Res. 13, 7264–7270 (2007).
Edwards, A.L. et al. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem. Biol. 20, 888–902 (2013).
Gavathiotis, E. et al. BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 (2008).
Leshchiner, E.S., Braun, C.R., Bird, G.H. & Walensky, L.D. Direct activation of full-length proapoptotic BAK. Proc. Natl. Acad. Sci. USA 110, E986–E995 (2013).
Malone, C.D. et al. Mcl-1 regulates the survival of adult neural precursor cells. Mol. Cell. Neurosci. 49, 439–447 (2012).
Opferman, J.T. et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307, 1101–1104 (2005).
Opferman, J.T. et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426, 671–676 (2003).
Wang, X. et al. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev. 27, 1351–1364 (2013).
Lim, S.M. et al. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew. Chem. Int. Edn. Engl. 53, 199–204 (2014).
Nussinov, R. & Tsai, C.J. The design of covalent allosteric drugs. Annu. Rev. Pharmacol. Toxicol. 55, 249–267 (2015).
Ge, Y. et al. Discovery and synthesis of hydronaphthoquinones as novel proteasome inhibitors. J. Med. Chem. 55, 1978–1998 (2012).
Barclay, L.A. et al. Inhibition of pro-apoptotic BAX by a noncanonical interaction mechanism. Mol. Cell 57, 873–886 (2015).
Braun, C.R. et al. Photoreactive stapled BH3 peptides to dissect the BCL-2 family interactome. Chem. Biol. 17, 1325–1333 (2010).
Wales, T.E. & Engen, J.R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006).
Kim, H. et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8, 1348–1358 (2006).
Liu, G. et al. High-quality NMR structure of human anti-apoptotic protein domain Mcl-1(171-327) for cancer drug design. PLoS One 9, e96521 (2014).
Sastry, G.M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 (2013).
Shelley, J.C. et al. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 21, 681–691 (2007).
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W. & Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Jorgensen, W.L., Maxwell, D.S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Hoover, W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 31, 1695–1697 (1985).
Martyna, G.J., Tobias, D.J. & Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Humphreys, D.D., Friesner, R.A. & Berne, B.J. A multiple-time-step molecular dynamics algorithm for macromoloecules. J. Phys. Chem. 98, 6885–6892 (1994).
Guo, Z. et al. Probing the alpha-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem. Biol. Drug Des. 75, 348–359 (2010).
Acknowledgements
We thank E. Smith for graphics support; J. Opferman (St. Jude Children's Research Hospital) for WT and Mcl1−/− MEFs; and W. Massefski (Dana-Farber Cancer Institute) for technical assistance with MAIM1 characterization by NMR. This research was supported by NIH grant 1R35CA197583, a Leukemia and Lymphoma Society (LLS) Marshall A. Lichtman Specialized Center of Research project grant, the Todd J. Schwartz Memorial Fund, the Wolpoff Family Foundation and an LLS Scholar Award (L.D.W.); NIH grant R01GM101135 and a Waters Corporation research collaboration (J.R.E.); and NIH grant T32GM007753 (J.L.).
Author information
Authors and Affiliations
Contributions
S.L., T.E.W., N.A.C., J.R.E. and L.D.W. designed the study; G.H.B. generated stapled peptides; D.T.C. synthesized and characterized MAIM1; C.G.G., D.T.C., S.E., A.J.H. and N.A.C. performed biochemical experiments; S.L. and T.E.W. conducted the HXMS experiments under the guidance of J.R.E.; S.E. and D.T.C. performed the cellular experiments; J.L. conducted molecular dynamics simulations; and S.L., T.E.W., S.E., D.T.C., J.L., J.R.E. and L.D.W. analyzed the data and wrote the manuscript, which was reviewed by all coauthors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Small-molecule covalent inhibition of MCL-1ΔNΔC.
(a) Fluorescence polarization dilutional binding assay of FITC−BID BH3 and MCL-1ΔNΔC in the presence and absence of N-(4-hydroxy-1-naphthalenyl)-benzenesulfonamide (CAS# 36942-42-4), a small molecule screening hit that exhibited irreversible inhibition of the BH3-binding activity of MCL-1ΔNΔC. (b) Chemical structures of MAIM1 analogs that emerged from our competitive MCL-1 SAHB A −MCL-1ΔNΔC screen. Competitive potency corresponded to compound reactivity, based on the nature of the leaving group (R1) and electron withdrawing capacity (R2). Data are mean ± s.e.m. (n = 3 technical replicates). (c) Chemical synthesis of MAIM1. (d-e) Trypsin digestion and LC-MS/MS of unmodified (d) and MAIM1-conjugated (e) MCL-1ΔNΔC. Full scan spectra (left) demonstrate the peptide containing unmodified C286 (mass: 2329.18 Da) and the MAIM1 conjugate (mass: 2483.19 Da), reflecting the added mass of cleaved MAIM1 adduct (i.e. minus tosyl). MS/MS spectra (right) further confirm MAIM1 conjugation at position C286. Whereas the masses of ions y3, y4, y5, y6, and y12 are shared between the peptides, there is a discrete mass shift for ion y15, which contains C286. For clarity, only the y ions are labeled.
Supplementary Figure 2 C286S mutagenesis abrogates the MAIM1 effect on deuterium exchange of MCL-1ΔNΔC in the presence of BID BH3.
Relative difference plots of the relative deuterium incorporation of MCL-1ΔNΔC C286S in the presence of BID BH3 minus the relative deuterium incorporation of MCL-1ΔNΔC C286S (brown), and the relative deuterium incorporation of MCL-1ΔNΔC C286S in the presence of MAIM1 and BID BH3 minus the relative deuterium incorporation of MCL-1ΔNΔC C286S in the presence of MAIM1 (orange). Changes in deuterium exchange above a stringent significant threshold of 0.8 Da are shaded light purple (0.8 – 2 Da), purple (2 – 3.5 Da), and dark purple (> 3.5 Da). HXMS data are for deuterium labeling at 10 seconds and represent the average of two independent experiments using distinct preparations of protein, protein conjugates, and peptide and protein mixtures.
Supplementary Figure 3 C286W mutation impairs MCL-1ΔNΔC protection of BID BH3 from deuterium exchange.
(a-d) MS spectra of BID BH3 peptide at (a) 0, (b) 10, (c) 20, and (d) 40 seconds for peptide alone (black) or the peptide combined with MCL-1ΔNΔC (red) or MCL-1ΔNΔC C286W (purple).
Supplementary Figure 4 Microscopic images of MEFs subjected to serum withdrawal.
The indicated MEFs were maintained in (a) full media or (b) serum-free media for 16 h, followed by phase contrast microscopic imaging. Exemplary apoptotic bodies are indicated by the black arrows. Bar, 25 μm.
Supplementary Figure 5 C286S mutagenesis does not affect the cell-viability response to serum withdrawal of MCL-1-reconstituted MEFs.
(a) Western blot of lysates from Mcl-1-/- MEFs reconstituted with comparable levels of human, full-length, wild-type or C16S,C286S mutant MCL-1. See also Supplementary Data Set 1. (b) Cell viability (48 h) for the indicated MEFs subjected to serum withdrawal. Data are normalized to the corresponding MEFs maintained in complete media. Error bars are mean ± s.d. (n = 3 technical replicates). *, p <0.01 by two-tailed student’s t test; n.s., not statistically significant.
Supplementary Figure 6 Comparison of human and mouse structures of MCL-1 in the C286 and F267 region.
(a-c) An overlay of the unliganded human (grey, PDB ID 2MHS) and mouse (blue, PDB ID 1WSX) forms of MCL-1 demonstrate a similar structural disposition of C286 and F267 (a), with C286 (orange) pointing toward the surface (b) and F267 (green) juxtaposed to a confluence of internal hydrophobic residues (yellow) (c). (d-f) In contrast, an overlay of BH3-liganded forms of human (grey, PDB ID 2PQK) and mouse (blue, PDB ID 2ROC) MCL-1 demonstrate differential orientations of C286 and F267 (d), with the location of C286 (orange) shifted within a relatively more unfolded region of the protein but still pointing toward the surface (e), and F267 (green) slightly torqued but with similar orientation and adjacency to internal hydrophobic residues (yellow) (f).
Supplementary Figure 7 Comparative BH3-based pulldown of MCL-1 and MCL-1 C286W from reconstituted Mcl1–/– MEFs.
(a-b) Western blots of streptavidin pull-downs from lysates of the indicated MEFs treated with biotinylated MCL-1 SAHB D , a selective stapled BH3 inhibitor of MCL-1 (left). See also Supplementary Data Set 1. The relative amount of precipitated MCL-1 vs. MCL-1 C286W was quantitated by densitometry of immunoreactive MCL-1 bands in the pull-down as ratioed to input control (right). Two independent streptavidin pull-down (SAP) experiments from treated lysates of the MEF cultures are shown.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1377 kb)
Supplementary Data Set 1
MCL-1 and actin western blot analyses (PDF 194 kb)
MCL-1
Molecular dynamics simulation of wild-type MCL-1 (MOV 13053 kb)
MCL-1–MAIM1
Molecular dynamics simulation of MAIM1-derivatized MCL-1 (MOV 12245 kb)
MCL-1 C286W
Molecular dynamics simulation of C286W-mutant MCL-1 (MOV 11396 kb)
Rights and permissions
About this article
Cite this article
Lee, S., Wales, T., Escudero, S. et al. Allosteric inhibition of antiapoptotic MCL-1. Nat Struct Mol Biol 23, 600–607 (2016). https://doi.org/10.1038/nsmb.3223
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3223
This article is cited by
-
A redox switch regulates the structure and function of anti-apoptotic BFL-1
Nature Structural & Molecular Biology (2020)
-
Small-molecule allosteric inhibitors of BAX
Nature Chemical Biology (2019)
-
Photocatalytic proximity labelling of MCL-1 by a BH3 ligand
Communications Chemistry (2019)
-
Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments
Nature Methods (2019)
-
Allosteric sensitization of proapoptotic BAX
Nature Chemical Biology (2017)